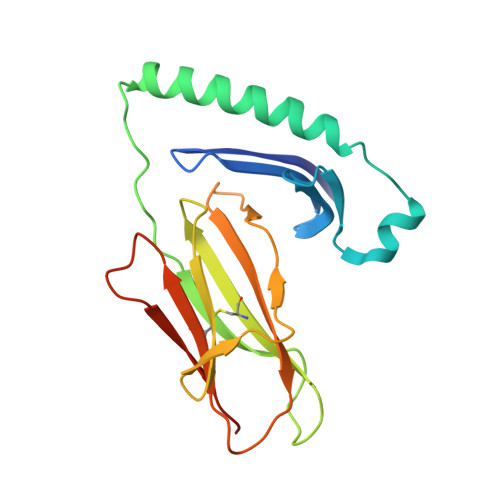
The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
Ting, Y.T., Petersen, J., Ramarathinam, S.H., Scally, S.W., Loh, K.L., Thomas, R., Suri, A., Baker, D.G., Purcell, A.W., Reid, H.H., Rossjohn, J.(2018) J Biological Chem 293: 3236-3251
- PubMed: 29317506
- DOI: https://doi.org/10.1074/jbc.RA117.001013
- Primary Citation of Related Structures:
6BIJ, 6BIL, 6BIN, 6BIR, 6BIV, 6BIX, 6BIY, 6BIZ - PubMed Abstract:
The HLA-DRB1 locus is strongly associated with rheumatoid arthritis (RA) susceptibility, whereupon citrullinated self-peptides bind to HLA-DR molecules bearing the shared epitope (SE) amino acid motif. However, the differing propensity for citrullinated/non-citrullinated self-peptides to bind given HLA-DR allomorphs remains unclear. Here, we used a fluorescence polarization assay to determine a hierarchy of binding affinities of 34 self-peptides implicated in RA against three HLA-DRB1 allomorphs (HLA-DRB1*04:01/*04:04/*04:05) each possessing the SE motif. For all three HLA-DRB1 allomorphs, we observed a strong correlation between binding affinity and citrullination at P4 of the bound peptide ligand. A differing hierarchy of peptide-binding affinities across the three HLA-DRB1 allomorphs was attributable to the β-chain polymorphisms that resided outside the SE motif and were consistent with sequences of naturally presented peptide ligands. Structural determination of eight HLA-DR4-self-epitope complexes revealed strict conformational convergence of the P4-Cit and surrounding HLA β-chain residues. Polymorphic residues that form part of the P1 and P9 pockets of the HLA-DR molecules provided a structural basis for the preferential binding of the citrullinated self-peptides to the HLA-DR4 allomorphs. Collectively, we provide a molecular basis for the interplay between citrullination of self-antigens and HLA polymorphisms that shape peptide-HLA-DR4 binding affinities in RA.
- From the Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute Monash University, and.
Organizational Affiliation: