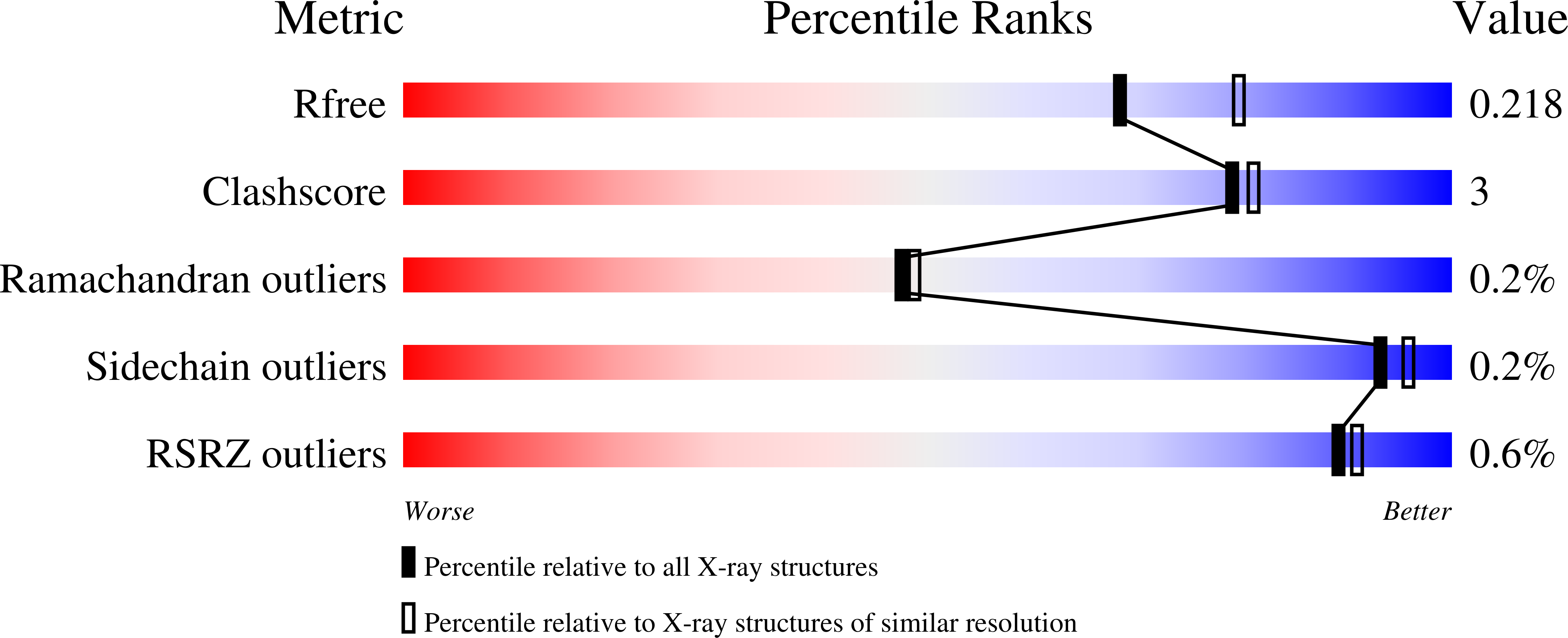
Human gamma-Glutamyl Transpeptidase 1: STRUCTURES OF THE FREE ENZYME, INHIBITOR-BOUND TETRAHEDRAL TRANSITION STATES, AND GLUTAMATE-BOUND ENZYME REVEAL NOVEL MOVEMENT WITHIN THE ACTIVE SITE DURING CATALYSIS.
Terzyan, S.S., Burgett, A.W., Heroux, A., Smith, C.A., Mooers, B.H., Hanigan, M.H.(2015) J Biological Chem 290: 17576-17586
- PubMed: 26013825
- DOI: https://doi.org/10.1074/jbc.M115.659680
- Primary Citation of Related Structures:
4Z9O, 4ZBK, 4ZC6, 4ZCG - PubMed Abstract:
γ-Glutamyl transpeptidase 1 (GGT1) is a cell surface, N-terminal nucleophile hydrolase that cleaves glutathione and other γ-glutamyl compounds. GGT1 expression is essential in cysteine homeostasis, and its induction has been implicated in the pathology of asthma, reperfusion injury, and cancer. In this study, we report four new crystal structures of human GGT1 (hGGT1) that show conformational changes within the active site as the enzyme progresses from the free enzyme to inhibitor-bound tetrahedral transition states and finally to the glutamate-bound structure prior to the release of this final product of the reaction. The structure of the apoenzyme shows flexibility within the active site. The serine-borate-bound hGGT1 crystal structure demonstrates that serine-borate occupies the active site of the enzyme, resulting in an enzyme-inhibitor complex that replicates the enzyme's tetrahedral intermediate/transition state. The structure of GGsTop-bound hGGT1 reveals its interactions with the enzyme and why neutral phosphonate diesters are more potent inhibitors than monoanionic phosphonates. These structures are the first structures for any eukaryotic GGT that include a molecule in the active site covalently bound to the catalytic Thr-381. The glutamate-bound structure shows the conformation of the enzyme prior to release of the final product and reveals novel information regarding the displacement of the main chain atoms that form the oxyanion hole and movement of the lid loop region when the active site is occupied. These data provide new insights into the mechanism of hGGT1-catalyzed reactions and will be invaluable in the development of new classes of hGGT1 inhibitors for therapeutic use.
- From the Macromolecular Crystallography Laboratory, Department of Biochemistry and Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma 73104.
Organizational Affiliation: