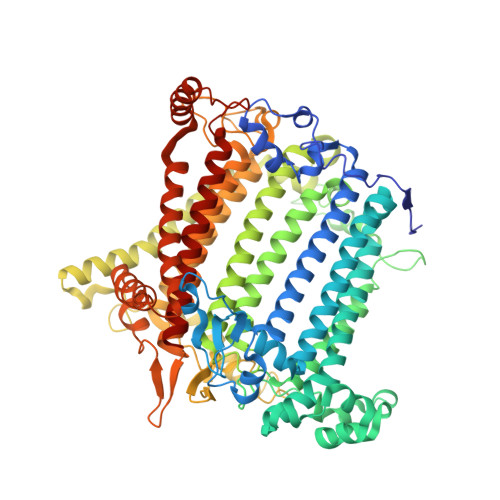
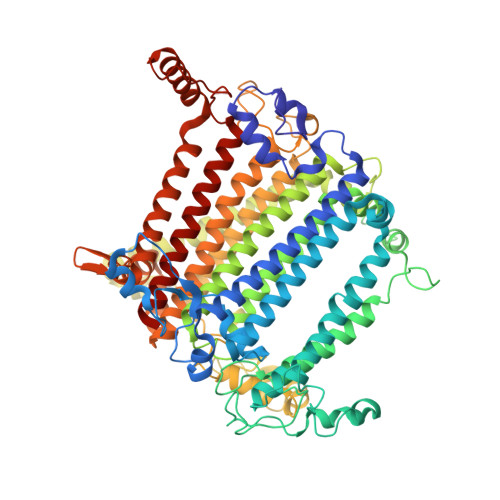
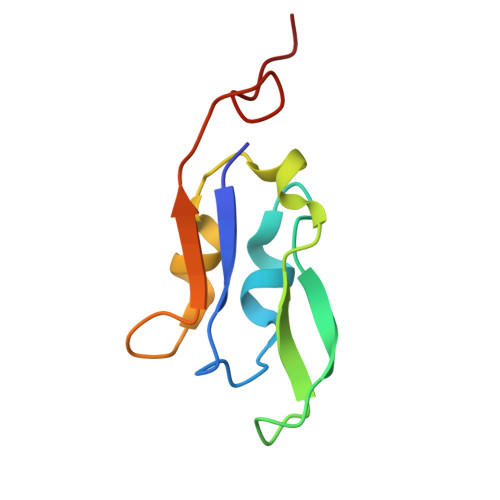
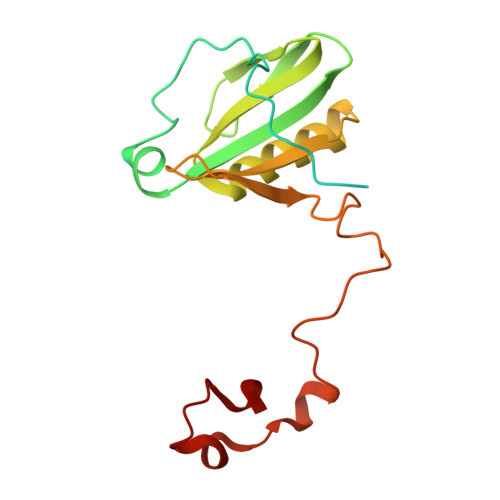
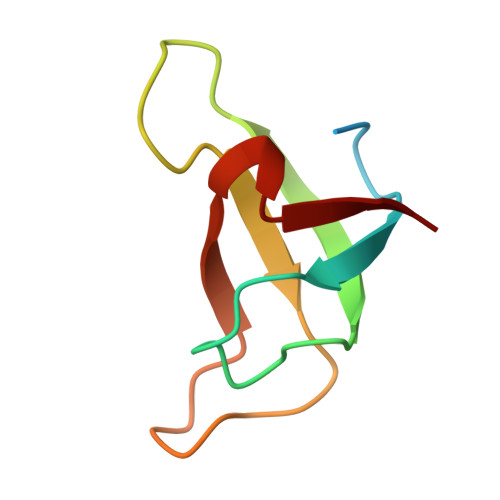
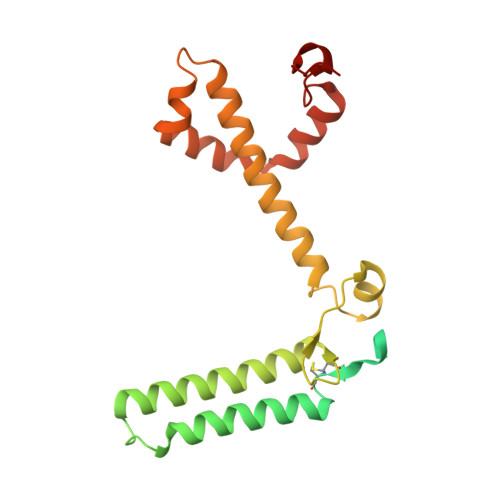
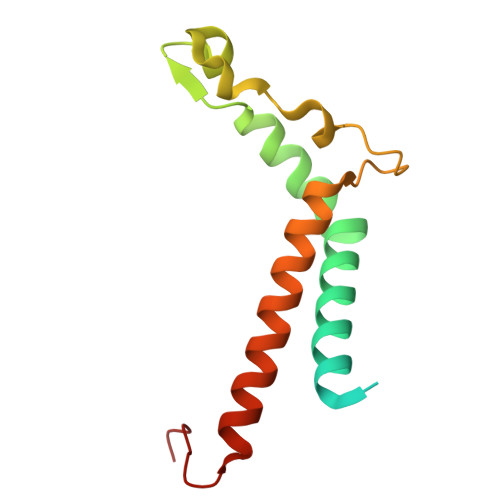
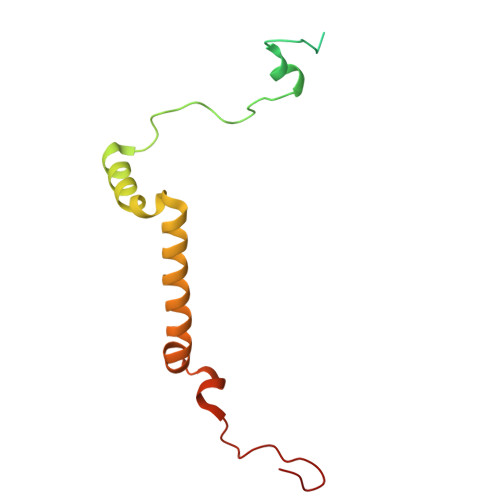
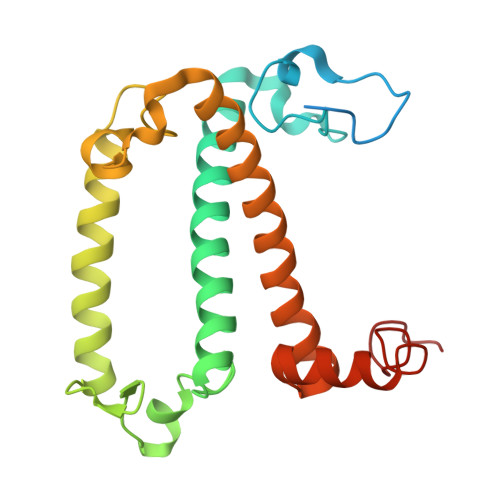
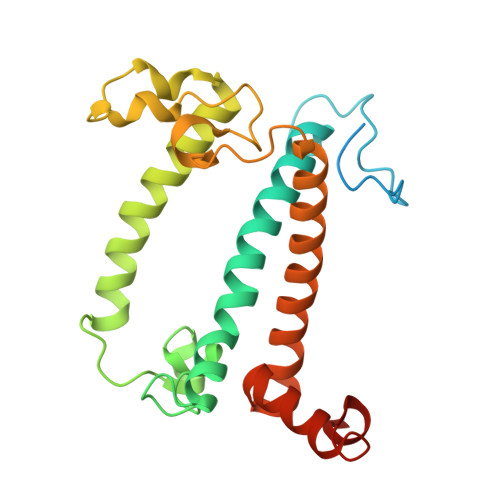
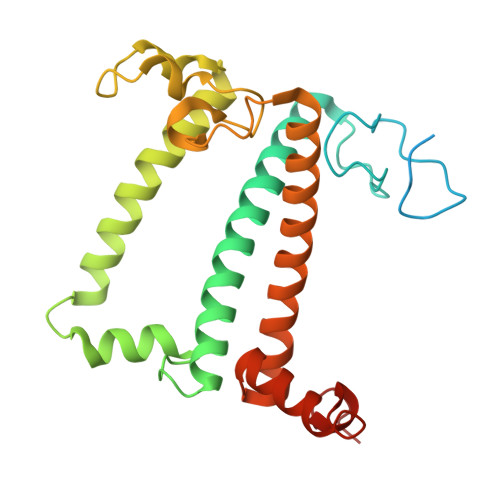
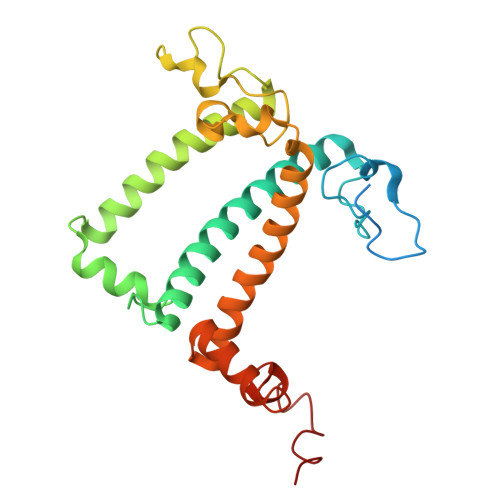
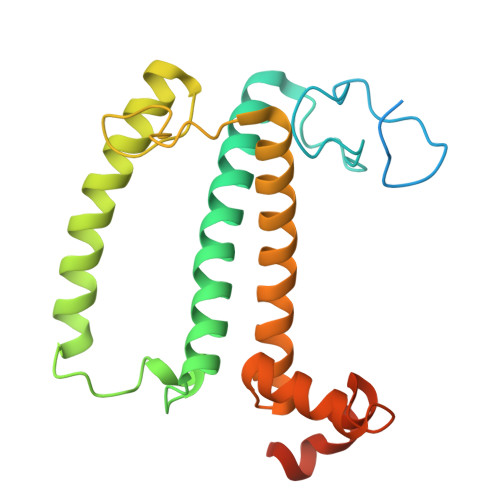
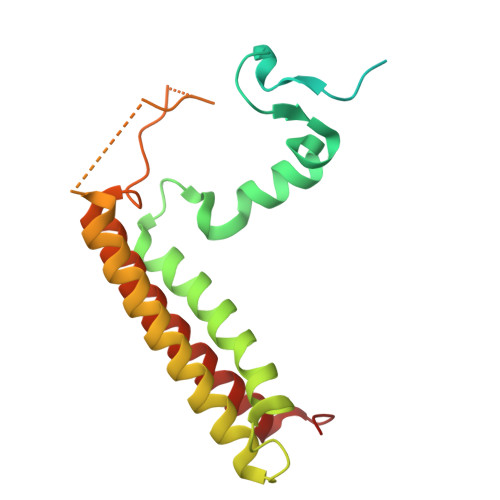
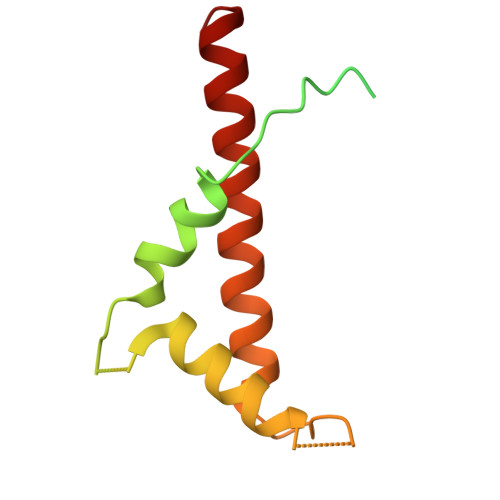
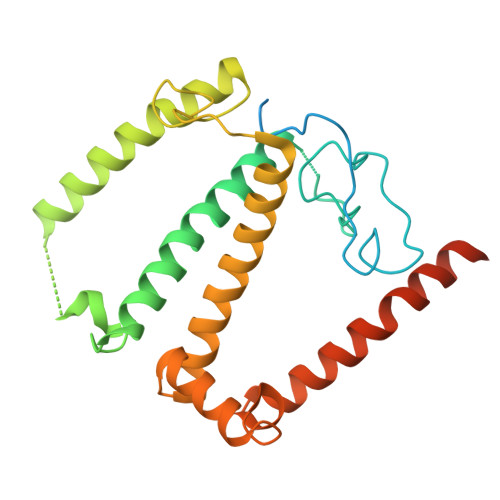
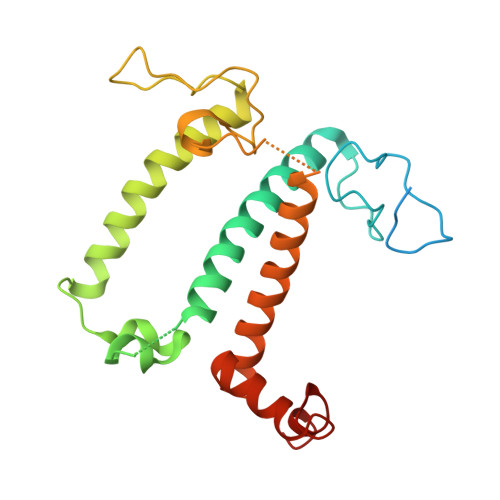
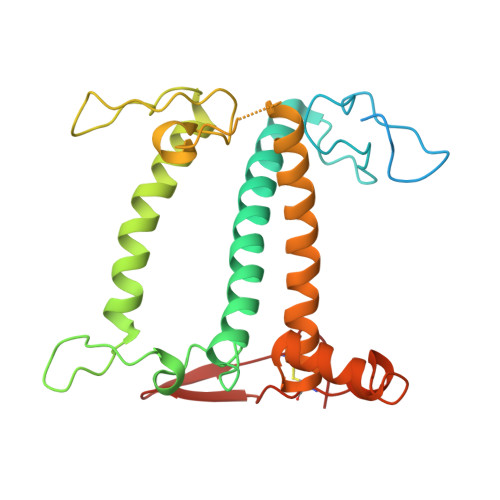
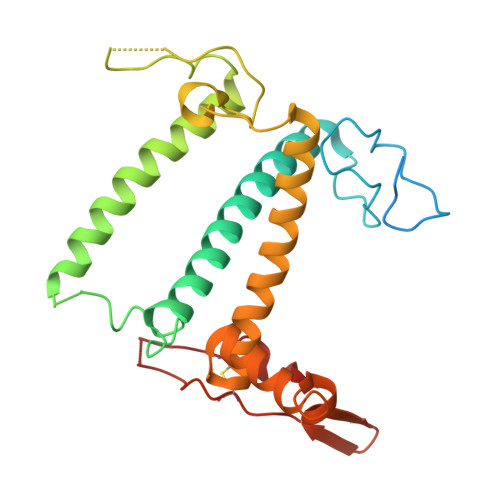
Structural study of the chlorophyll between Lhca8 and PsaJ in an Antarctica green algal photosystem I-LHCI supercomplex revealed by its atomic structure.
Tsai, P.C., Kato, K., Shen, J.R., Akita, F.(2025) Biochim Biophys Acta Bioenerg 1866: 149543-149543
- PubMed: 39947506
- DOI: https://doi.org/10.1016/j.bbabio.2025.149543
- Primary Citation of Related Structures:
9KQP, 9KQQ - PubMed Abstract:
Coccomyxa subellipsoidea is an oleaginous, non-motile unicellular green microalga isolated from Antarctica, and is an attractive candidate for CO 2 fixation and biomass production. C. subellipsoidea is the first polar green alga whose genome has been sequenced. Understanding the structure of photosystems from C. subellipsoidea can provide more information about the conversion of light energy into chemical energy under extreme environments. Photosystems I (PSI) is one of the two photosystems highly conserved from cyanobacteria to vascular plants, and associates with a large amount of outer light-harvesting complex (LHC) which absorb light energy and transfer them to the core complex. Here, we determined the structure of the PSI-10 LHCIs and PSI-8 LHCIs supercomplexes from C. subellipsoidea at 1.92 Å and 2.06 Å resolutions by cryo-electron microscopy, respectively. The supercomplex is similar to PSI-LHCI from other green algae, whereas a large amount of water molecules is observed in our structure because of the high-resolution map. Two novel chlorophylls (Chls), Chl a321 in Lhca4 and Chl a314 in Lhca8, are observed at the lumenal side in our structure, in which Lhca8-Chl a314 provides a potential excitation energy transfer (EET) pathway between the inner-belt of LHCI and the core at the lumenal side. A total of three major EET pathways from LHCIs to PSI core are proposed, and C. subellipsoidea might adapt to the extreme environment by transferring energy in these three different EET pathways instead of by two major pathways proposed in other organisms.
- Research Institute for Interdisciplinary Science, and Graduate School of Environmental, Life, Natural Science and Technology, Okayama University, Okayama 700-8530, Japan.
Organizational Affiliation: