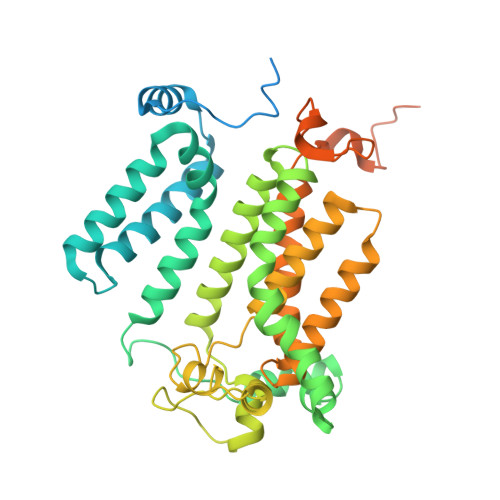
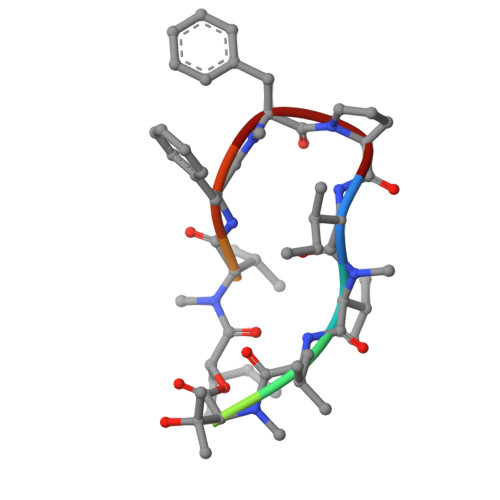
Mechanisms of aureobasidin A inhibition and drug resistance in a fungal IPC synthase complex.
Wu, X., Gong, X., Xie, T.(2025) Nat Commun 16: 5010-5010
- PubMed: 40442105
- DOI: https://doi.org/10.1038/s41467-025-60423-y
- Primary Citation of Related Structures:
9IVC - PubMed Abstract:
The enzyme inositol phosphorylceramide (IPC) synthase is essential for survival and virulence in fungi, while absent in mammals, thus representing a potential target for antifungal treatments. Aureobasidin A (AbA), a natural cyclic peptide, displays antifungal activity and inhibits IPC synthase, but the precise molecular mechanism remains unclear. Here, we present the cryo-EM structure of the Saccharomyces cerevisiae IPC synthase, composed of catalytic subunit Aur1 and regulatory subunit Kei1, in its AbA-bound state. The complex is resolved as a dimer of Aur1-Kei1 heterodimers, with Aur1 mediating homodimerization. AbA occupies a predominantly hydrophobic pocket in the catalytic core domain of each Aur1 subunit, blocking the entry of both substrates. Mutations conferring AbA resistance cluster near the AbA-binding site, thus interfering with AbA binding. Our study lays a foundation for the development of therapeutic drugs targeting fungal IPC synthase.
- Shenzhen Key Laboratory of Plant Genetic Engineering and Molecular Design, Department of Biology, School of Life Sciences, Southern University of Science and Technology, Shenzhen, Guangdong, 518055, China.
Organizational Affiliation: