Structure-Based Analysis of Semisynthetic Anti-TB Rufomycin Analogues.
Zhou, B., Shetye, G., Klein, L.L., Wolf, N.M., Lee, H., McAlpine, J.B., Harris, G., Chen, S.N., Suh, J.W., Cho, S.H., Franzblau, S.G., Abad-Zapatero, C., Pauli, G.F.(2025) J Nat Prod 88: 907-925
- PubMed: 40126472
- DOI: https://doi.org/10.1021/acs.jnatprod.4c01266
- Primary Citation of Related Structures:
9DIN - PubMed Abstract:
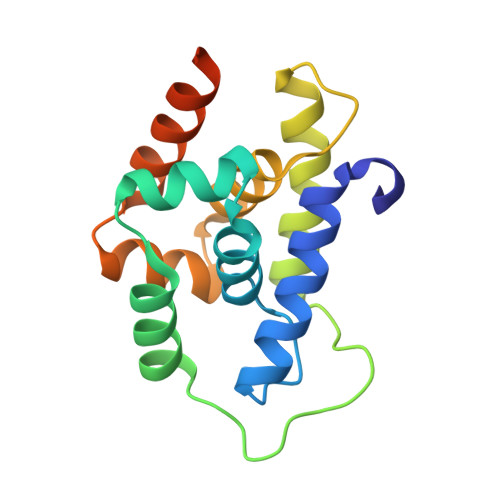
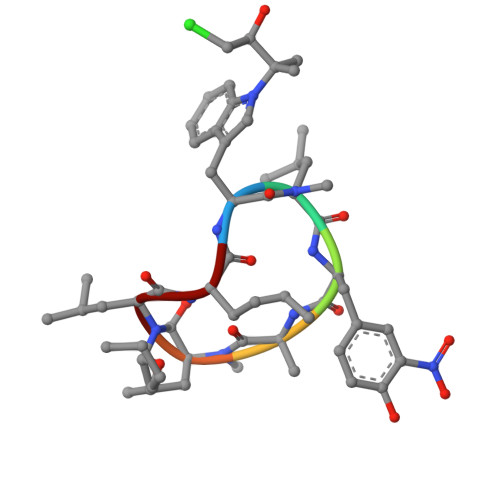
This study employed structural information from cocrystals of rufomycin 4 ( 1a ) and caseinolytic protein C1 (ClpC1)-NTD-wt to guide design and semisynthesis of rufomycin analogues, evaluate their antituberculosis (TB) biological profiles, and establish structure-activity relationships (SAR). Covering three regions of interest (ROIs, A-C) as modification sites, 14 of the 30 semisynthetic analogues ( 2 - 31 ) showed similar or improved MICs relative to the main natural precursors, rufomycins 4/6 ( 1a/b ). Compounds 5 and 27 exhibited up to 10-fold enhanced potency against Mycobacterium tuberculosis ( Mtb ) in vitro, with MIC values of 1.9 and 1.4 nM, respectively. Evaluation of ClpC1-binding properties used existing ClpC1-NTD complexes with rufomycin 4 (PDB: 6cn8) and ecumicin (PDB: 6pbs) as references. The newly reported X-ray ClpC1-NTD cocrystal structure of 11 (syn. But4-Cl) revealed significant conformational effects involving the side chains of certain amino acids of the heptapeptide and confirmed the importance of ROIs A-C for medicinal chemistry efforts. Observed interactions of the N -terminal tail of ClpC1 with the rufomycin analogues vs ecumicin explains their different modes of inactivating the ClpC1/P1/P2 homeostatic machinery. Collectively, the observations inform further SAR optimization strategies for the rufomycin class of antibiotics and complement our understanding of their mode of action.
- Myongji Bioefficacy Research Center, Myongji University, Myongji-Ro 116, Yongin, Gyeonggi-Do 17058, Republic of Korea.
Organizational Affiliation: