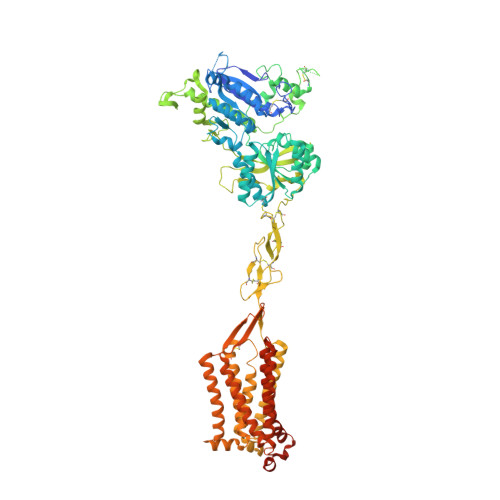Promiscuous G-protein activation by the calcium-sensing receptor.
Zuo, H., Park, J., Frangaj, A., Ye, J., Lu, G., Manning, J.J., Asher, W.B., Lu, Z., Hu, G.B., Wang, L., Mendez, J., Eng, E., Zhang, Z., Lin, X., Grassucci, R., Hendrickson, W.A., Clarke, O.B., Javitch, J.A., Conigrave, A.D., Fan, Q.R.(2024) Nature 629: 481-488
- PubMed: 38632411
- DOI: https://doi.org/10.1038/s41586-024-07331-1
- Primary Citation of Related Structures:
9ASB, 9AVG, 9AVL, 9AXF, 9AYF - PubMed Abstract:
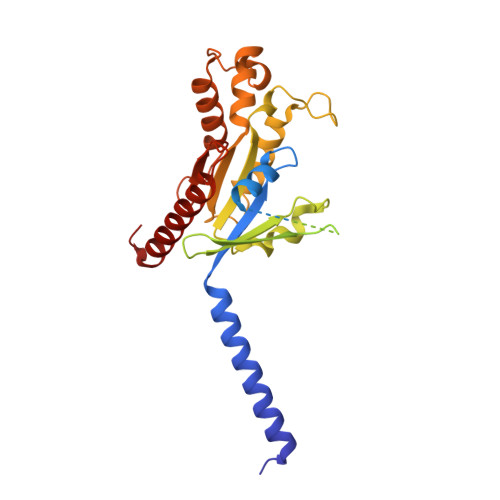
The human calcium-sensing receptor (CaSR) detects fluctuations in the extracellular Ca 2+ concentration and maintains Ca 2+ homeostasis 1,2 . It also mediates diverse cellular processes not associated with Ca 2+ balance 3-5 . The functional pleiotropy of CaSR arises in part from its ability to signal through several G-protein subtypes 6 . We determined structures of CaSR in complex with G proteins from three different subfamilies: G q , G i and G s . We found that the homodimeric CaSR of each complex couples to a single G protein through a common mode. This involves the C-terminal helix of each Gα subunit binding to a shallow pocket that is formed in one CaSR subunit by all three intracellular loops (ICL1-ICL3), an extended transmembrane helix 3 and an ordered C-terminal region. G-protein binding expands the transmembrane dimer interface, which is further stabilized by phospholipid. The restraint imposed by the receptor dimer, in combination with ICL2, enables G-protein activation by facilitating conformational transition of Gα. We identified a single Gα residue that determines G q and G s versus G i selectivity. The length and flexibility of ICL2 allows CaSR to bind all three Gα subtypes, thereby conferring capacity for promiscuous G-protein coupling.
- Department of Molecular Pharmacology and Therapeutics, Columbia University, New York, NY, USA.
Organizational Affiliation: