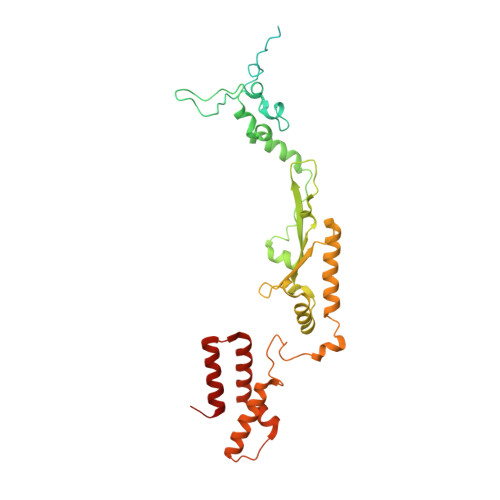
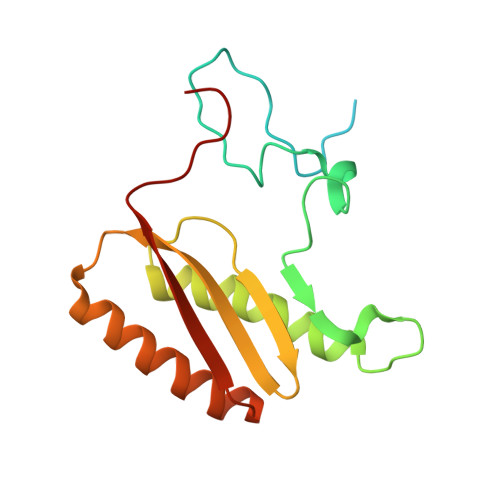
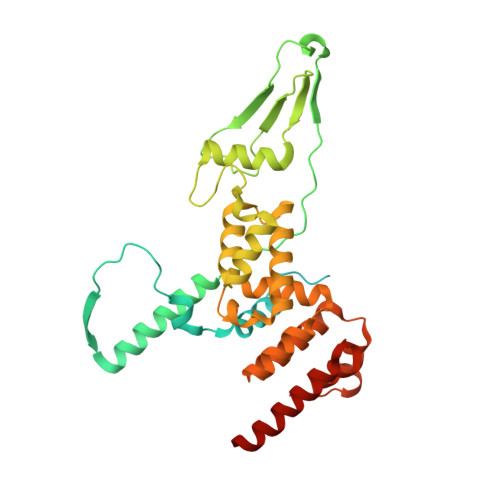
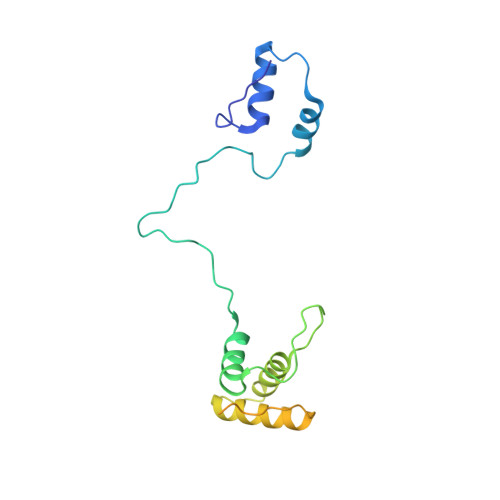
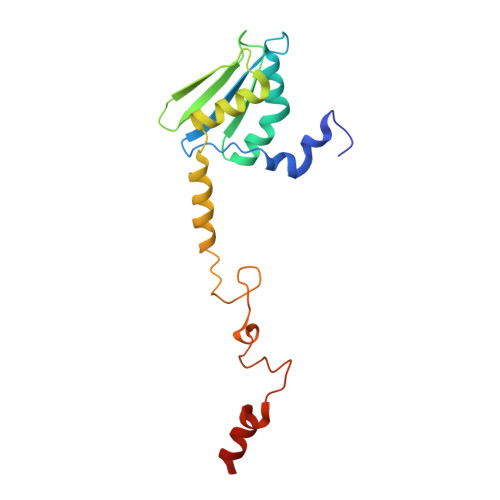
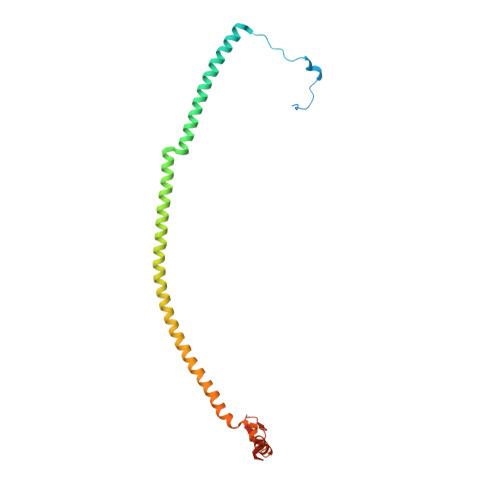
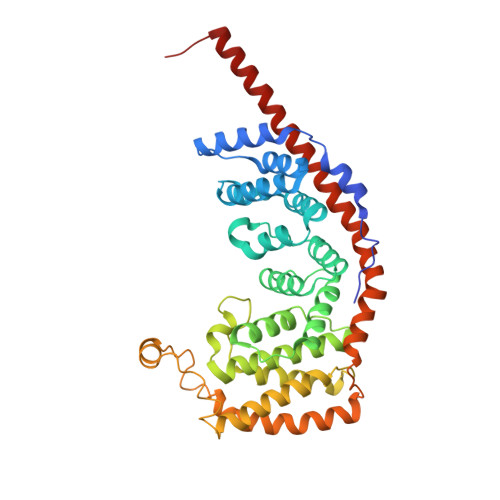
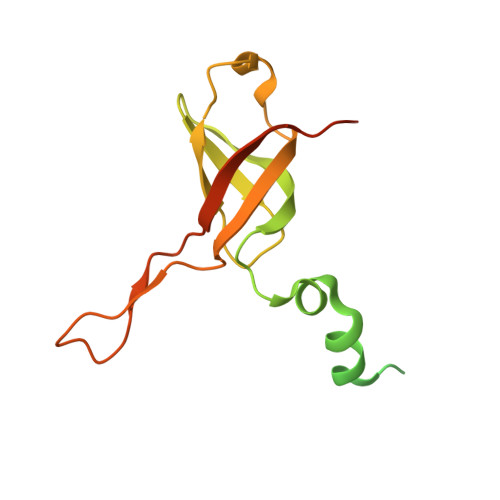
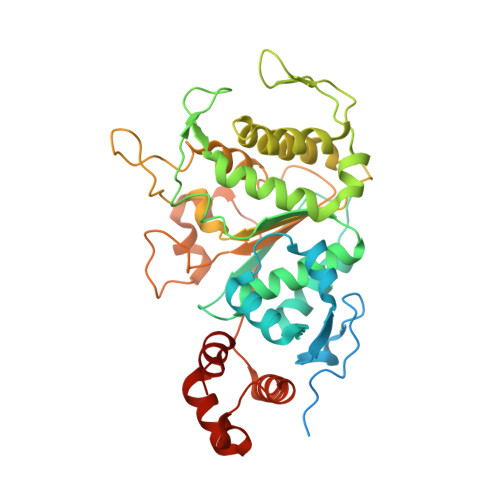
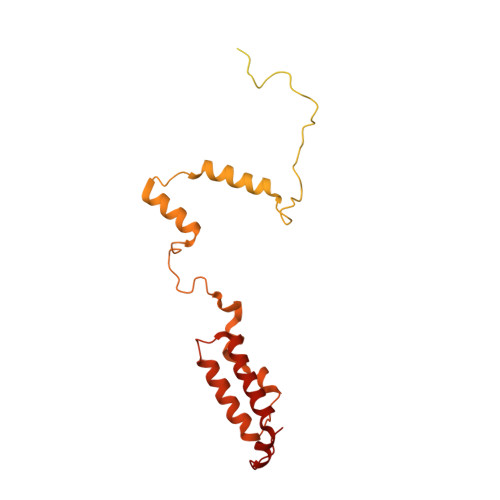
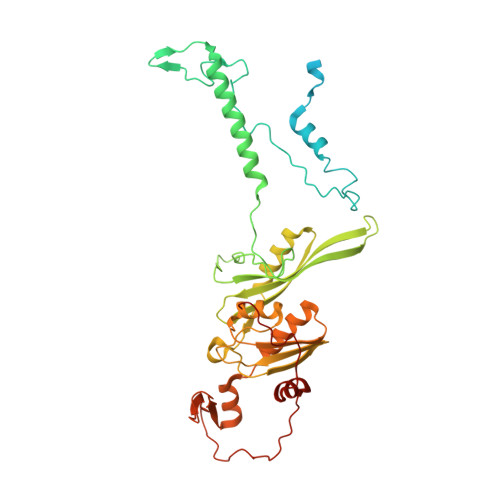
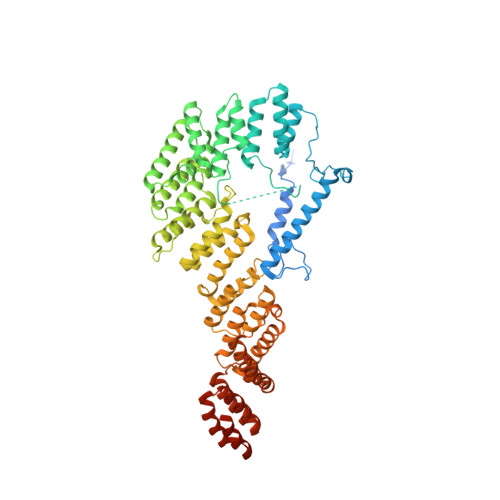
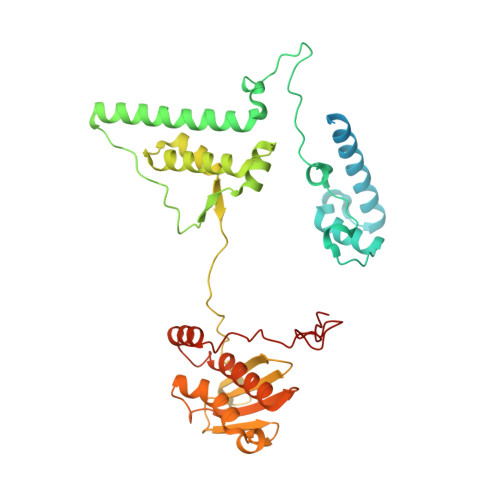
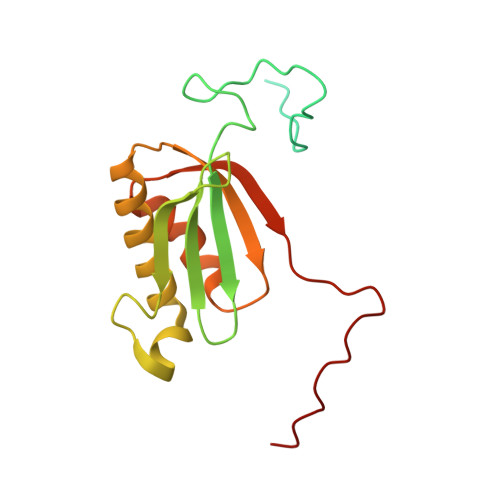
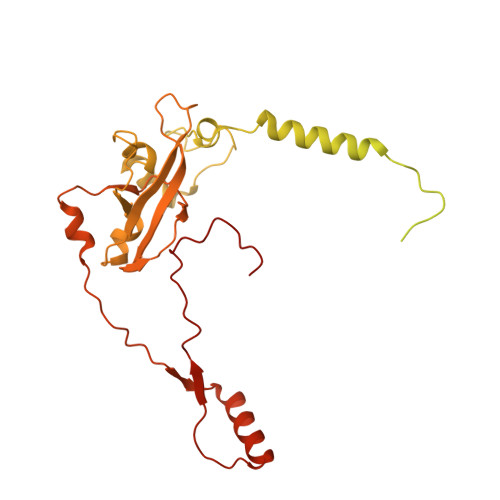
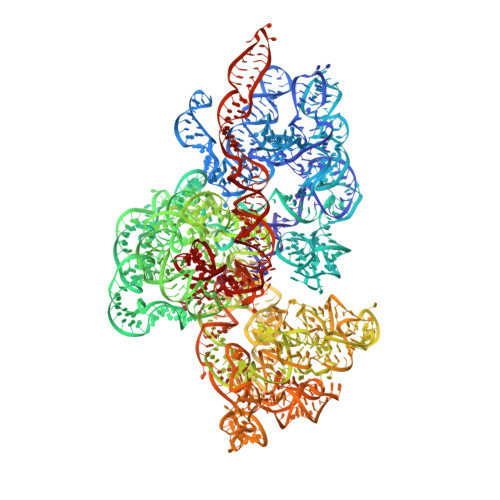
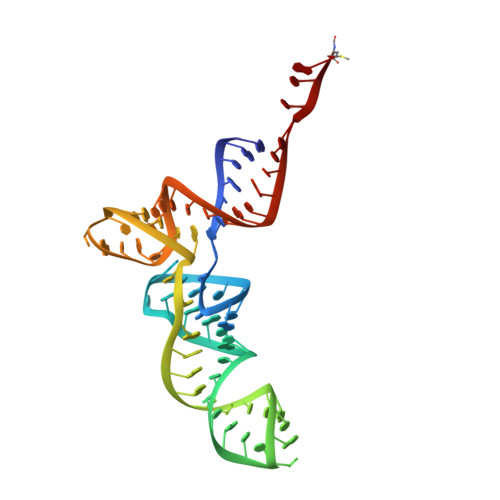
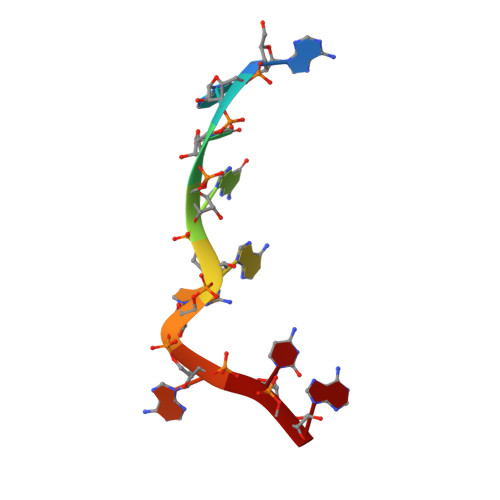
Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Saurer, M., Leibundgut, M., Nadimpalli, H.P., Scaiola, A., Schonhut, T., Lee, R.G., Siira, S.J., Rackham, O., Dreos, R., Lenarcic, T., Kummer, E., Gatfield, D., Filipovska, A., Ban, N.(2023) Science 380: 531-536
- PubMed: 37141370
- DOI: https://doi.org/10.1126/science.adf9890
- Primary Citation of Related Structures:
8OIN, 8OIP, 8OIQ, 8OIR, 8OIS, 8OIT - PubMed Abstract:
The genetic code that specifies the identity of amino acids incorporated into proteins during protein synthesis is almost universally conserved. Mitochondrial genomes feature deviations from the standard genetic code, including the reassignment of two arginine codons to stop codons. The protein required for translation termination at these noncanonical stop codons to release the newly synthesized polypeptides is not currently known. In this study, we used gene editing and ribosomal profiling in combination with cryo-electron microscopy to establish that mitochondrial release factor 1 (mtRF1) detects noncanonical stop codons in human mitochondria by a previously unknown mechanism of codon recognition. We discovered that binding of mtRF1 to the decoding center of the ribosome stabilizes a highly unusual conformation in the messenger RNA in which the ribosomal RNA participates in specific recognition of the noncanonical stop codons.
- Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zürich, 8093 Zürich, Switzerland.
Organizational Affiliation: