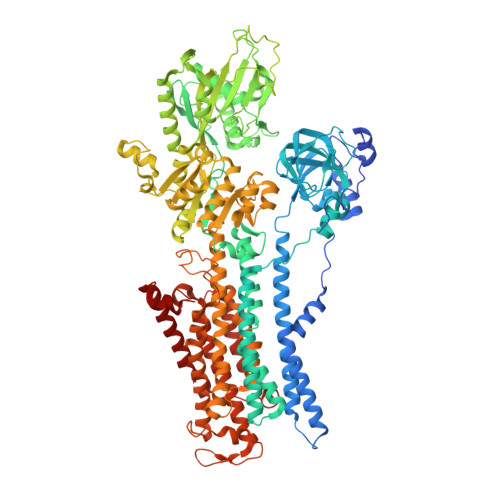
Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Kanai, R., Vilsen, B., Cornelius, F., Toyoshima, C.(2023) FEBS Lett 597: 1957-1976
- PubMed: 37357620
- DOI: https://doi.org/10.1002/1873-3468.14689
- Primary Citation of Related Structures:
8JBK, 8JBL, 8JBM, 8JFZ - PubMed Abstract:
Na + ,K + -ATPase (NKA) plays a pivotal role in establishing electrochemical gradients for Na + and K + across the cell membrane by alternating between the E1 (showing high affinity for Na + and low affinity for K + ) and E2 (low affinity to Na + and high affinity to K + ) forms. Presented here are two crystal structures of NKA in E1·Mg 2+ and E1·3Na + states at 2.9 and 2.8 Å resolution, respectively. These two E1 structures fill a gap in our description of the NKA reaction cycle based on the atomic structures. We describe how NKA converts the K + -bound E2·2K + form to an E1 (E1·Mg 2+ ) form, which allows high-affinity Na + binding, eventually closing the cytoplasmic gate (in E1 ~ P·ADP·3Na + ) after binding three Na + , while keeping the extracellular ion pathway sealed. We now understand previously unknown functional roles for several parts of NKA and that NKA uses even the lipid bilayer for gating the ion pathway.
- Institute for Quantitative Biosciences, The University of Tokyo, Bunkyo-ku, Japan.
Organizational Affiliation: