New crystal system to promote screening for new eukaryotic inhibitors
Kolosova, O., Zgadzay, Y., Wu, C., Validov, S., Usachev, K., Jenner, L., Sachs, M.S., Yusupova, G., Yusupov, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L2-B | D [auth j], EC [auth AW] | 254 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PF08 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PF08 Go to UniProtKB: A0A1D8PF08 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PF08 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L3 | E [auth k], FC [auth AX] | 389 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q59LS1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q59LS1 Go to UniProtKB: Q59LS1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q59LS1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L4-B | F [auth l], GC [auth AY] | 363 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PFV1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PFV1 Go to UniProtKB: A0A1D8PFV1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PFV1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized protein CaJ7.0206 | G [auth m], HC [auth AZ] | 298 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5AGZ7 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5AGZ7 Go to UniProtKB: Q5AGZ7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5AGZ7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L6 | H [auth n], IC [auth BA] | 176 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PCX8 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PCX8 Go to UniProtKB: A0A1D8PCX8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PCX8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L7-A | I [auth o], JC [auth BB] | 241 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PDL6 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PDL6 Go to UniProtKB: A0A1D8PDL6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PDL6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L8 | J [auth p], KC [auth BC] | 262 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5ANA1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5ANA1 Go to UniProtKB: Q5ANA1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5ANA1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L9-B | K [auth q], LC [auth BD] | 191 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5AEN2 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5AEN2 Go to UniProtKB: Q5AEN2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5AEN2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L10 | L [auth r], MC [auth BE] | 220 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5AIB8 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5AIB8 Go to UniProtKB: Q5AIB8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5AIB8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L11-B | M [auth s], NC [auth BF] | 174 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PHW1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PHW1 Go to UniProtKB: A0A1D8PHW1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PHW1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L13 | N [auth t], OC [auth BG] | 202 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for O59931 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore O59931 Go to UniProtKB: O59931 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O59931 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L14-B | O [auth u], PC [auth BH] | 131 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PFL9 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PFL9 Go to UniProtKB: A0A1D8PFL9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PFL9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L15-A | P [auth v], QC [auth BI] | 204 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5A6R1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5A6R1 Go to UniProtKB: Q5A6R1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5A6R1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein L13 | Q [auth w], RC [auth BJ] | 200 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5AB87 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5AB87 Go to UniProtKB: Q5AB87 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5AB87 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein L22 | R [auth x], SC [auth BK] | 185 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q59TE0 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q59TE0 Go to UniProtKB: Q59TE0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q59TE0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 19 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L18-A | S [auth y], TC [auth BL] | 186 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK43 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK43 Go to UniProtKB: A0A1D8PK43 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK43 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 20 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L19-A | T [auth z], UC [auth BM] | 190 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK40 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK40 Go to UniProtKB: A0A1D8PK40 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK40 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 21 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L20 | U [auth 0], VC [auth BN] | 172 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PLC9 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PLC9 Go to UniProtKB: A0A1D8PLC9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PLC9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 22 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L21-A | V [auth 2], WC [auth BO] | 160 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PGY0 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PGY0 Go to UniProtKB: A0A1D8PGY0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PGY0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 23 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L22-B | W [auth 5], XC [auth BP] | 124 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PM41 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PM41 Go to UniProtKB: A0A1D8PM41 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PM41 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 24 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L23-A | X [auth 6], YC [auth BQ] | 137 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PPT5 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PPT5 Go to UniProtKB: A0A1D8PPT5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PPT5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 25 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L24-A | Y [auth 7], ZC [auth BR] | 155 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5A6A1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5A6A1 Go to UniProtKB: Q5A6A1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5A6A1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 26 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L25 | AD [auth BS], Z [auth 8] | 142 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PPS1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PPS1 Go to UniProtKB: A0A1D8PPS1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PPS1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 27 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein L24 | AA [auth 9], BD [auth BT] | 127 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PCQ5 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PCQ5 Go to UniProtKB: A0A1D8PCQ5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PCQ5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 28 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L27 | BA [auth AA], CD [auth BU] | 136 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9P843 (Candida albicans) Explore Q9P843 Go to UniProtKB: Q9P843 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9P843 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 29 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L28 | CA [auth AB], DD [auth BV] | 149 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PSC5 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PSC5 Go to UniProtKB: A0A1D8PSC5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PSC5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 30 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L29 | DA [auth AC], ED [auth BW] | 63 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PF57 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PF57 Go to UniProtKB: A0A1D8PF57 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PF57 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 31 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L30 | EA [auth AD], FD [auth BX] | 106 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PM75 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PM75 Go to UniProtKB: A0A1D8PM75 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PM75 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 32 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L31-B | FA [auth AE], GD [auth BY] | 112 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F1M1 (Candida albicans) Explore A0A8H6F1M1 Go to UniProtKB: A0A8H6F1M1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F1M1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 33 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L32 | GA [auth AF], HD [auth BZ] | 131 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PPN6 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PPN6 Go to UniProtKB: A0A1D8PPN6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PPN6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 34 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L33-A | HA [auth AG], ID [auth CA] | 107 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PHH4 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PHH4 Go to UniProtKB: A0A1D8PHH4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PHH4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 35 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L34-B | IA [auth AH], JD [auth CB] | 122 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PDZ1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PDZ1 Go to UniProtKB: A0A1D8PDZ1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PDZ1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 36 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein L29 | JA [auth AI], KD [auth CC] | 120 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK30 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK30 Go to UniProtKB: A0A1D8PK30 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK30 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 37 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L36 | KA [auth AJ], LD [auth CD] | 99 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PH21 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PH21 Go to UniProtKB: A0A1D8PH21 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PH21 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 38 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L37-B | LA [auth AK], MD [auth CE] | 87 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PF45 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PF45 Go to UniProtKB: A0A1D8PF45 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PF45 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 39 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L38 | MA [auth AL], ND [auth CF] | 78 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PG16 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PG16 Go to UniProtKB: A0A1D8PG16 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PG16 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 40 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L39 | NA [auth AM], OD [auth CG] | 51 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PDT4 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PDT4 Go to UniProtKB: A0A1D8PDT4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PDT4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 41 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L40-B | OA [auth AN], PD [auth CH] | 52 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PL68 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PL68 Go to UniProtKB: A0A1D8PL68 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PL68 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 42 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L41 | PA [auth AO], QD [auth CI] | 25 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P9WER7 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore P9WER7 Go to UniProtKB: P9WER7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WER7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 43 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L42-B | QA [auth AP], RD [auth CJ] | 106 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PEV4 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PEV4 Go to UniProtKB: A0A1D8PEV4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PEV4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 44 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L43-A | RA [auth AQ], SD [auth CK] | 92 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PP14 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PP14 Go to UniProtKB: A0A1D8PP14 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PP14 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 45 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein CAALFM_C304810CA | SA [auth i], TD [auth CL] | 267 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK71 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK71 Go to UniProtKB: A0A1D8PK71 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK71 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 47 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S0 | UA [auth C], VD [auth CN] | 261 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for O42817 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore O42817 Go to UniProtKB: O42817 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O42817 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 48 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S1 | VA [auth D], WD [auth CO] | 256 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P40910 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore P40910 Go to UniProtKB: P40910 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P40910 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 49 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S5 | WA [auth E], XD [auth CP] | 249 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5A900 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5A900 Go to UniProtKB: Q5A900 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5A900 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 50 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S3 | XA [auth F], YD [auth CQ] | 251 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PSV5 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PSV5 Go to UniProtKB: A0A1D8PSV5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PSV5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 51 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S4 | YA [auth G], ZD [auth CR] | 262 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PCI6 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PCI6 Go to UniProtKB: A0A1D8PCI6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PCI6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 52 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S7 | AE [auth CS], ZA [auth H] | 225 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5AG43 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5AG43 Go to UniProtKB: Q5AG43 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5AG43 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 53 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S6 | AB [auth I], BE [auth CT] | 236 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PL99 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PL99 Go to UniProtKB: A0A1D8PL99 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PL99 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 54 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S7 | BB [auth J], CE [auth CU] | 186 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5AJ93 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5AJ93 Go to UniProtKB: Q5AJ93 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5AJ93 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 55 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S8 | CB [auth K], DE [auth CV] | 206 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q59T44 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q59T44 Go to UniProtKB: Q59T44 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q59T44 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 56 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S4 | DB [auth L], EE [auth CW] | 189 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PGY8 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PGY8 Go to UniProtKB: A0A1D8PGY8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PGY8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 57 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S10-A | EB [auth M], FE [auth CX] | 118 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PI15 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PI15 Go to UniProtKB: A0A1D8PI15 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PI15 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 58 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S11A | FB [auth N], GE [auth CY] | 155 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PN83 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PN83 Go to UniProtKB: A0A1D8PN83 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PN83 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 59 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S12 | GB [auth O], HE [auth CZ] | 143 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5ADQ6 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5ADQ6 Go to UniProtKB: Q5ADQ6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5ADQ6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 60 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S13 | HB [auth P], IE [auth DA] | 151 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PPE0 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PPE0 Go to UniProtKB: A0A1D8PPE0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PPE0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 61 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S14-A | IB [auth Q], JE [auth DB] | 132 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PDT3 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PDT3 Go to UniProtKB: A0A1D8PDT3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PDT3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 62 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S15 | JB [auth R], KE [auth DC] | 142 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK22 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK22 Go to UniProtKB: A0A1D8PK22 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK22 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 63 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S16 | KB [auth S], LE [auth DD] | 142 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PCW6 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PCW6 Go to UniProtKB: A0A1D8PCW6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PCW6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 64 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S17-B | LB [auth T], ME [auth DE] | 137 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PEY9 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PEY9 Go to UniProtKB: A0A1D8PEY9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PEY9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 65 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S18-B | MB [auth U], NE [auth DF] | 145 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PQQ5 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PQQ5 Go to UniProtKB: A0A1D8PQQ5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PQQ5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 66 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S19-A | NB [auth V], OE [auth DG] | 145 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK61 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK61 Go to UniProtKB: A0A1D8PK61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 67 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S10 | OB [auth W], PE [auth DH] | 119 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5A389 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5A389 Go to UniProtKB: Q5A389 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5A389 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 68 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S21 | PB [auth X], QE [auth DI] | 87 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PCG7 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PCG7 Go to UniProtKB: A0A1D8PCG7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PCG7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 69 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S22-A | QB [auth Y], RE [auth DJ] | 130 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q96W54 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q96W54 Go to UniProtKB: Q96W54 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96W54 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 70 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S23 (S12) | RB [auth Z], SE [auth DK] | 145 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PDU3 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PDU3 Go to UniProtKB: A0A1D8PDU3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PDU3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 71 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S24 | SB [auth a], TE [auth DL] | 135 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5A7K0 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5A7K0 Go to UniProtKB: Q5A7K0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5A7K0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 72 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S25 | TB [auth b], UE [auth DM] | 105 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PNQ6 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PNQ6 Go to UniProtKB: A0A1D8PNQ6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PNQ6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 73 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S26 | UB [auth c], VE [auth DN] | 119 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5ALV6 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5ALV6 Go to UniProtKB: Q5ALV6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5ALV6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 74 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S27 | VB [auth d], WE [auth DO] | 82 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PTI7 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PTI7 Go to UniProtKB: A0A1D8PTI7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PTI7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 75 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S28-B | WB [auth e], XE [auth DP] | 67 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PQN0 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PQN0 Go to UniProtKB: A0A1D8PQN0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PQN0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 76 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S29A | XB [auth f], YE [auth DQ] | 56 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PTR4 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PTR4 Go to UniProtKB: A0A1D8PTR4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PTR4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 77 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S30 | YB [auth g], ZE [auth DR] | 63 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PSK2 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PSK2 Go to UniProtKB: A0A1D8PSK2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PSK2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 78 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquitin-40S ribosomal protein S31 fusion protein | AF [auth DS], ZB [auth h] | 193 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5A109 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5A109 Go to UniProtKB: Q5A109 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5A109 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 79 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein subunit beta-like protein | AC [auth AR], BF [auth DT] | 317 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P83774 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore P83774 Go to UniProtKB: P83774 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P83774 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 80 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S acidic ribosomal protein P0 | CF [auth P0], GF [auth p0] | 312 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for G1UA71 (Candida albicans) Explore G1UA71 Go to UniProtKB: G1UA71 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G1UA71 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 81 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L12-A | DF [auth 12] | 165 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5AJF7 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5AJF7 Go to UniProtKB: Q5AJF7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5AJF7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 82 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein | EF [auth L1], FF [auth l1] | 217 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9UVJ4 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q9UVJ4 Go to UniProtKB: Q9UVJ4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9UVJ4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 25S | A [auth 1], BC [auth AS] | 3,359 | Candida albicans | 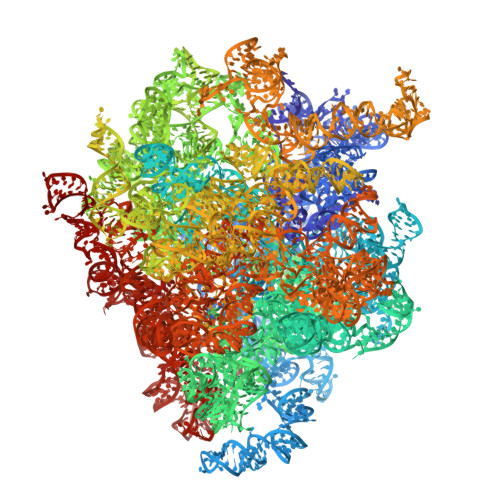 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5S | B [auth 3], CC [auth AT] | 121 | Candida albicans | 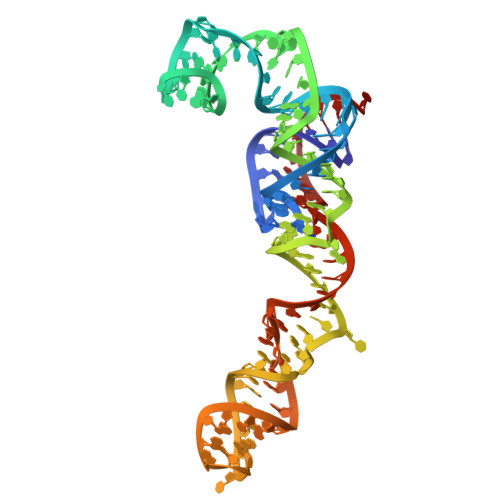 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5.8S | C [auth 4], DC [auth AU] | 158 | Candida albicans | 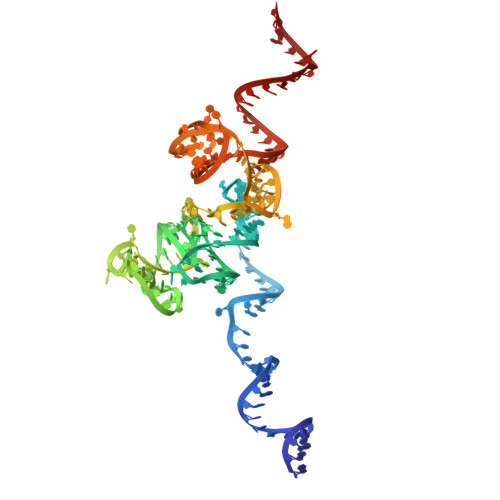 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 46 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 18S | TA [auth B], UD [auth CM] | 1,787 | Candida albicans | 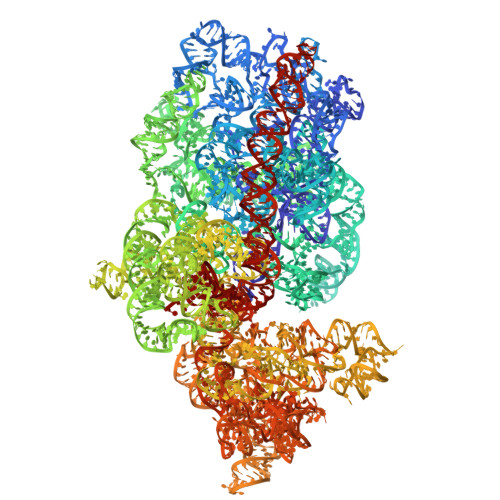 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 3K5 Query on 3K5 | JF [auth 1], OLA [auth AS] | 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose C40 H52 O17 VOTNXJVGRXZYOA-XFJWYURVSA-N |  | ||
| TIX Query on TIX | FAB [auth AS], SAA [auth 1] | ailanthone C20 H26 O7 DOVPXPXYKBQEAK-BJTBSANGSA-N |  | ||
| ZN Query on ZN | AEA [auth AQ] BJB [auth DQ] CJB [auth DS] HCB [auth CB] HLA [auth c] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | AAA [auth 1] AAB [auth AS] ABA [auth 3] ABB [auth AW] ACA [auth k] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 298.96 | α = 90 |
| b = 294.09 | β = 100.12 |
| c = 450.21 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Foundation for Medical Research (France) | France | DEQ20181039600 |
| Foundation for Medical Research (France) | France | DBF20160635745 |
| Foundation for Medical Research (France) | France | FDT202204014886 |
| Russian Science Foundation | Russian Federation | 20-65-47031 |
| National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health & Human Development (NIH/NICHD) | United States | 21 AI138158 |