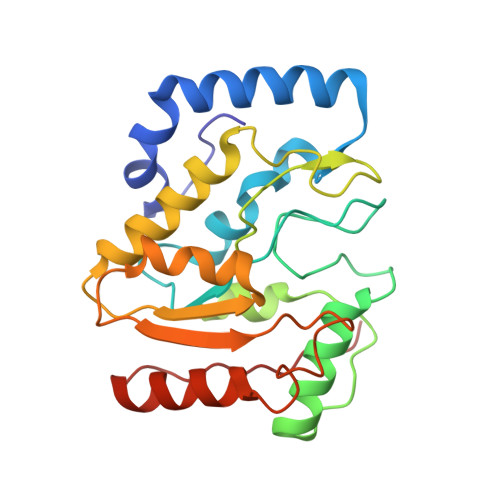
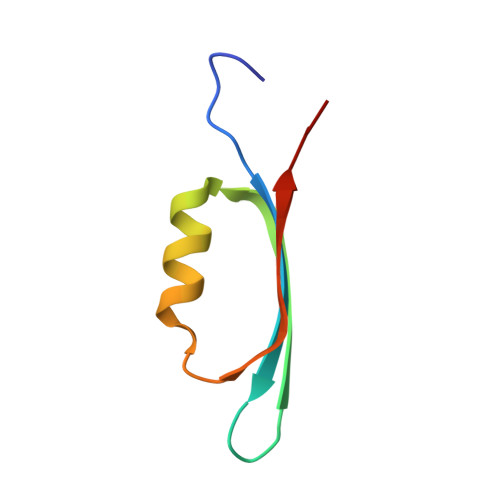
A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Muselmani, W., Kashif-Khan, N., Bagneris, C., Santangelo, R., Williams, M.A., Savva, R.(2023) Viruses 15
- PubMed: 37376646
- DOI: https://doi.org/10.3390/v15061348
- Primary Citation of Related Structures:
8AIL, 8AIM, 8AIN - PubMed Abstract:
DNA-mimicking proteins encoded by viruses can modulate processes such as innate cellular immunity. An example is Ung-family uracil-DNA glycosylase inhibition, which prevents Ung-mediated degradation via the stoichiometric protein blockade of the Ung DNA-binding cleft. This is significant where uracil-DNA is a key determinant in the replication and distribution of virus genomes. Unrelated protein folds support a common physicochemical spatial strategy for Ung inhibition, characterised by pronounced sequence plasticity within the diverse fold families. That, and the fact that relatively few template sequences are biochemically verified to encode Ung inhibitor proteins, presents a barrier to the straightforward identification of Ung inhibitors in genomic sequences. In this study, distant homologs of known Ung inhibitors were characterised via structural biology and structure prediction methods. A recombinant cellular survival assay and in vitro biochemical assay were used to screen distant variants and mutants to further explore tolerated sequence plasticity in motifs supporting Ung inhibition. The resulting validated sequence repertoire defines an expanded set of heuristic sequence and biophysical signatures shared by known Ung inhibitor proteins. A computational search of genome database sequences and the results of recombinant tests of selected output sequences obtained are presented here.
- Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, Malet Street, London WC1E 7HX, UK.
Organizational Affiliation: