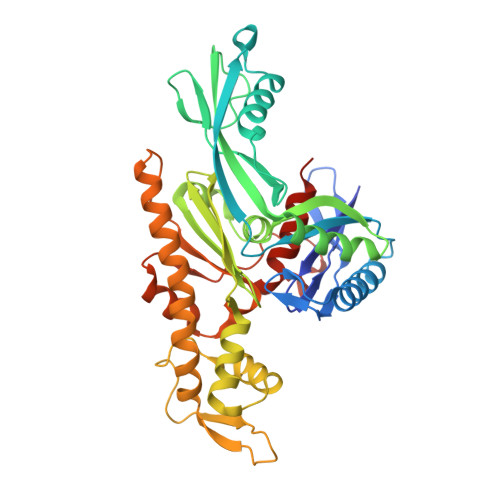
Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nierhaus, T., McLaughlin, S.H., Burmann, F., Kureisaite-Ciziene, D., Maslen, S.L., Skehel, J.M., Yu, C.W.H., Freund, S.M.V., Funke, L.F.H., Chin, J.W., Lowe, J.(2022) Nat Microbiol 7: 1686-1701
- PubMed: 36123441
- DOI: https://doi.org/10.1038/s41564-022-01206-9
- Primary Citation of Related Structures:
7Q6D, 7Q6F, 7Q6G, 7Q6I - PubMed Abstract:
During bacterial cell division, filaments of tubulin-like FtsZ form the Z-ring, which is the cytoplasmic scaffold for divisome assembly. In Escherichia coli, the actin homologue FtsA anchors the Z-ring to the membrane and recruits divisome components, including bitopic FtsN. FtsN regulates the periplasmic peptidoglycan synthase FtsWI. To characterize how FtsA regulates FtsN, we applied electron microscopy to show that E. coli FtsA forms antiparallel double filaments on lipid monolayers when bound to the cytoplasmic tail of FtsN. Using X-ray crystallography, we demonstrate that Vibrio maritimus FtsA crystallizes as an equivalent double filament. We identified an FtsA-FtsN interaction site in the IA-IC interdomain cleft of FtsA using X-ray crystallography and confirmed that FtsA forms double filaments in vivo by site-specific cysteine cross-linking. FtsA-FtsN double filaments reconstituted in or on liposomes prefer negative Gaussian curvature, like those of MreB, the actin-like protein of the elongasome. We propose that curved antiparallel FtsA double filaments together with treadmilling FtsZ filaments organize septal peptidoglycan synthesis in the division plane.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: