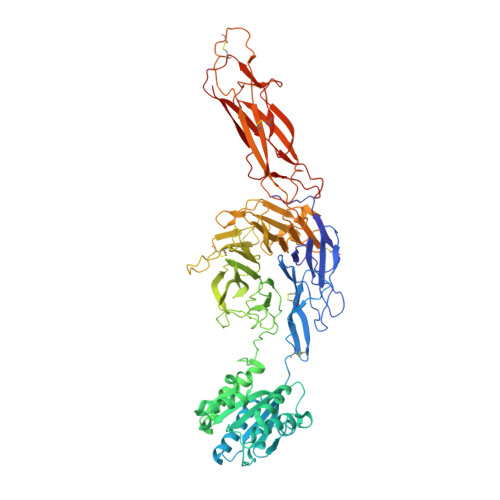
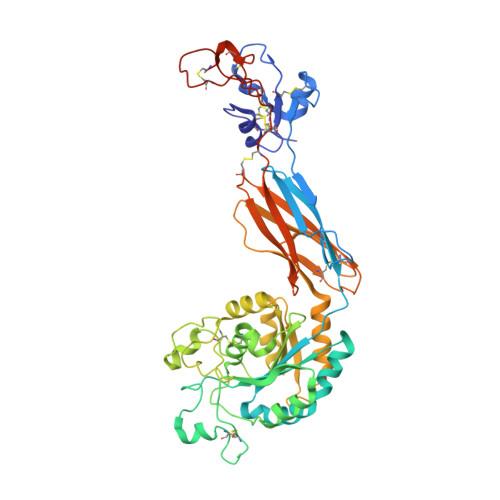
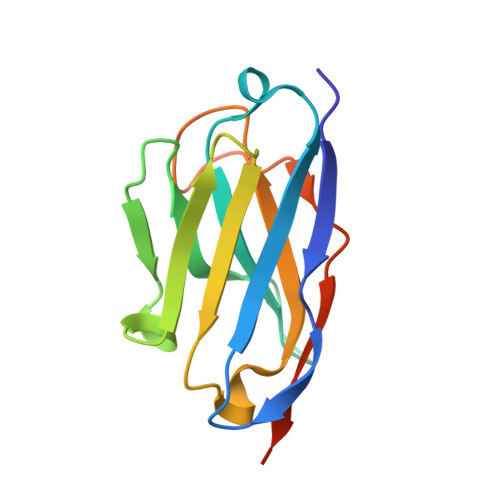
Structural insights into the function-modulating effects of nanobody binding to the integrin receptor alpha M beta 2.
Jensen, R.K., Pedersen, H., Lorentzen, J., Laursen, N.S., Vorup-Jensen, T., Andersen, G.R.(2022) J Biological Chem 298: 102168-102168
- PubMed: 35738398
- DOI: https://doi.org/10.1016/j.jbc.2022.102168
- Primary Citation of Related Structures:
7NP9, 7P2D - PubMed Abstract:
The integrin receptor α M β 2 mediates phagocytosis of complement-opsonized objects, adhesion to the extracellular matrix, and transendothelial migration of leukocytes. However, the mechanistic aspects of α M β 2 signaling upon ligand binding are unclear. Here, we present the first atomic structure of the human α M β 2 headpiece fragment in complex with the nanobody (Nb) hCD11bNb1 at a resolution of 3.2 Å. We show that the receptor headpiece adopts the closed conformation expected to exhibit low ligand affinity. The crystal structure indicates that in the R77H α M variant, associated with systemic lupus erythematosus, the modified allosteric relationship between ligand binding and integrin outside-inside signaling is due to subtle conformational effects transmitted over a distance of 40 Å. Furthermore, we found the Nb binds to the αI domain of the α M subunit in an Mg 2+ -independent manner with low nanomolar affinity. Biochemical and biophysical experiments with purified proteins demonstrated that the Nb acts as a competitive inhibitor through steric hindrance exerted on the thioester domain of complement component iC3b attempting to bind the α M subunit. Surprisingly, we show that the Nb stimulates the interaction of cell-bound α M β 2 with iC3b, suggesting that it may represent a novel high-affinity proteinaceous α M β 2 -specific agonist. Taken together, our data suggest that the iC3b-α M β 2 complex may be more dynamic than predicted from the crystal structure of the core complex. We propose a model based on the conformational spectrum of the receptor to reconcile these observations regarding the functional consequences of hCD11bNb1 binding to α M β 2 .
- Department of Molecular Biology and Genetics, Aarhus University, Denmark.
Organizational Affiliation: