Synthesis and evaluation of peptidic thrombin inhibitors bearing acid-stable sulfotyrosine analogues.
Dowman, L.J., Agten, S.M., Ripoll-Rozada, J., Calisto, B.M., Pereira, P.J.B., Payne, R.J.(2021) Chem Commun (Camb) 57: 10923-10926
- PubMed: 34596182
- DOI: https://doi.org/10.1039/d1cc04742f
- Primary Citation of Related Structures:
7PHX - PubMed Abstract:
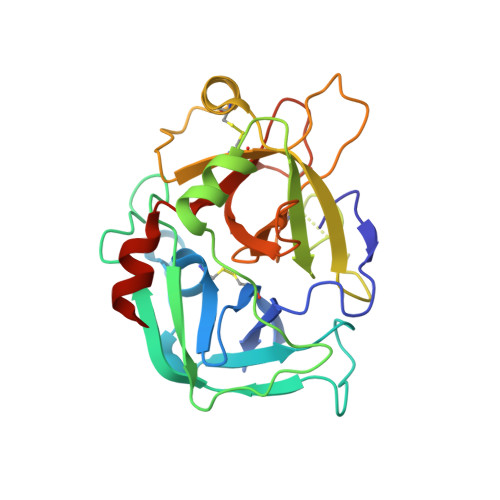
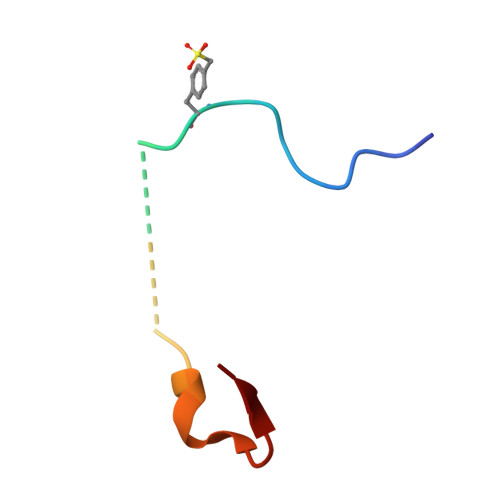
Tyrosine sulfation is an important post-translational modification of peptides and proteins which underpins and modulates many protein-protein interactions. In order to overcome the inherent instability of the native modification, we report the synthesis of two sulfonate analogues and their incorporation into two thrombin-inhibiting sulfopeptides. The effective mimicry of these sulfonate analogues for native sulfotyrosine was validated in the context of their thrombin inhibitory activity and binding mode, as determined by X-ray crystallography.
- School of Chemistry, The University of Sydney, Sydney, NSW 2006, Australia. richard.payne@sydney.edu.au.
Organizational Affiliation: