Structural insights into preinitiation complex assembly on core promoters.
Chen, X., Qi, Y., Wu, Z., Wang, X., Li, J., Zhao, D., Hou, H., Li, Y., Yu, Z., Liu, W., Wang, M., Ren, Y., Li, Z., Yang, H., Xu, Y.(2021) Science 372
- PubMed: 33795473
- DOI: https://doi.org/10.1126/science.aba8490
- Primary Citation of Related Structures:
7EDX, 7EG7, 7EG8, 7EG9, 7EGA, 7EGB, 7EGC, 7EGD, 7EGE, 7EGF, 7EGG, 7EGH, 7EGI, 7EGJ - PubMed Abstract:
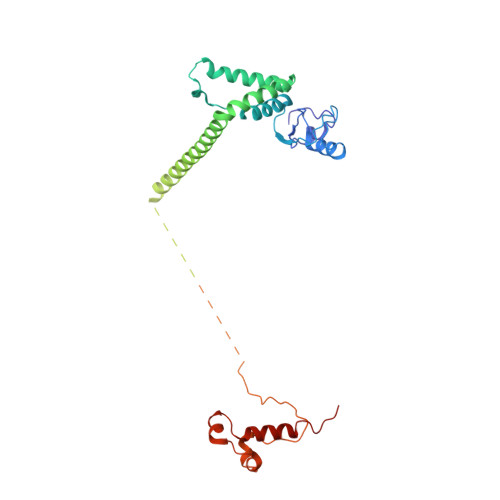
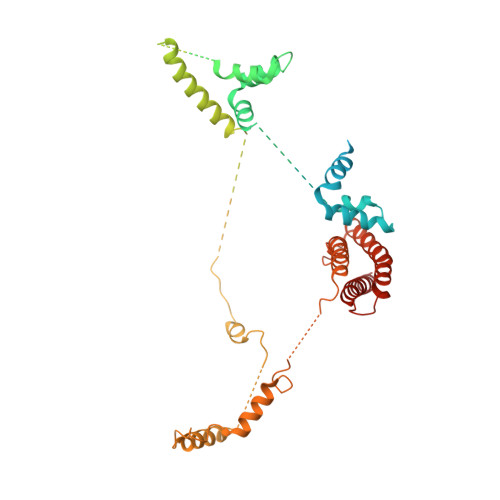
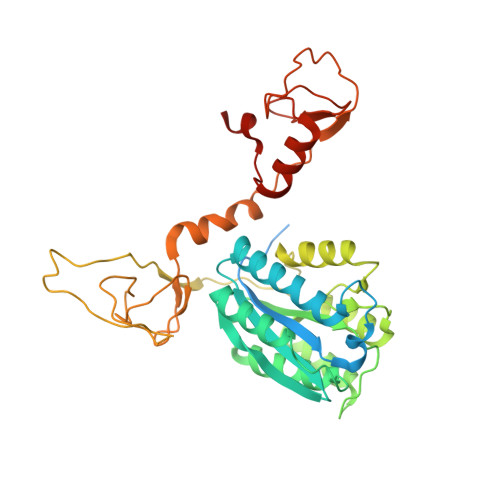
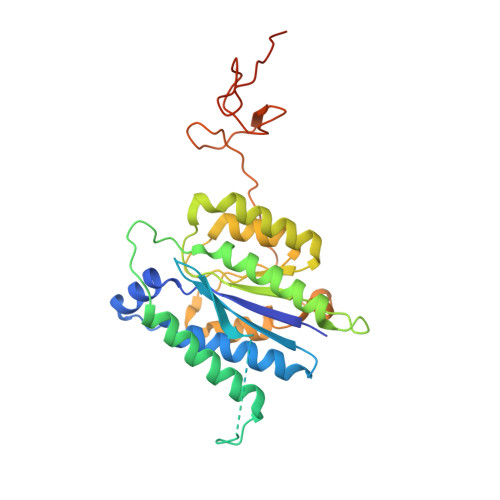
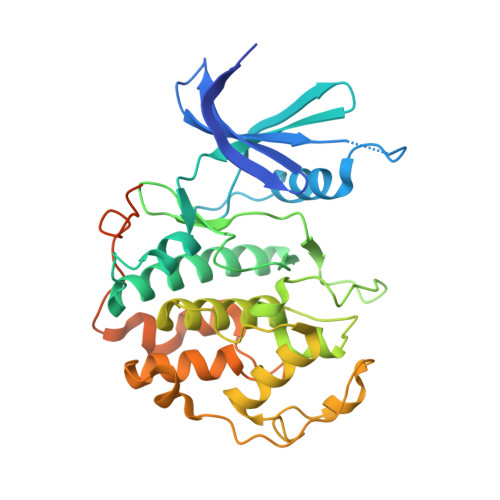
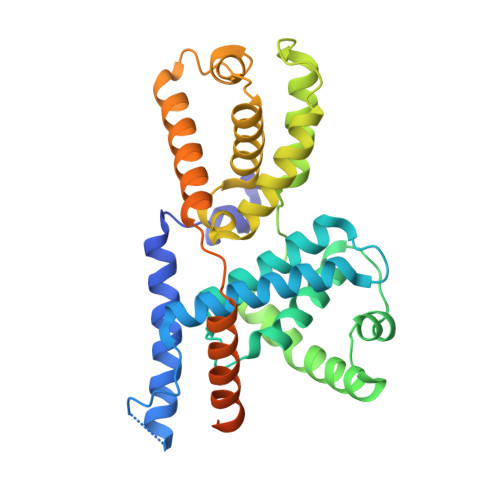
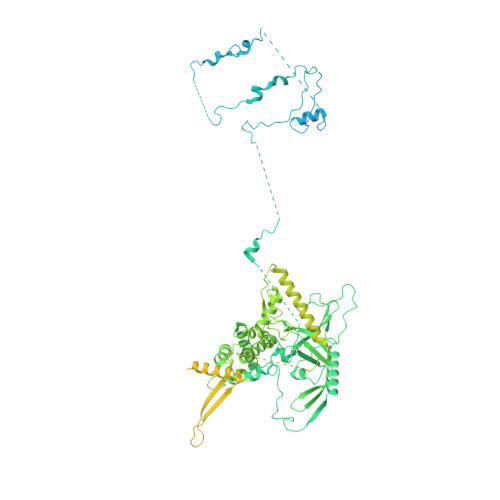
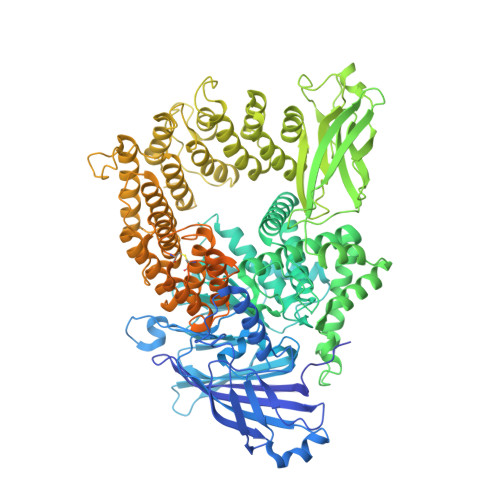
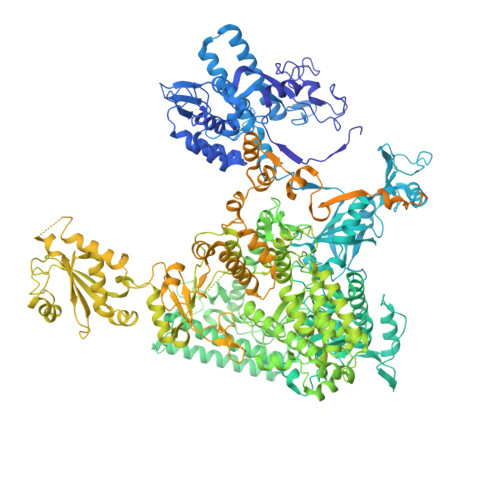
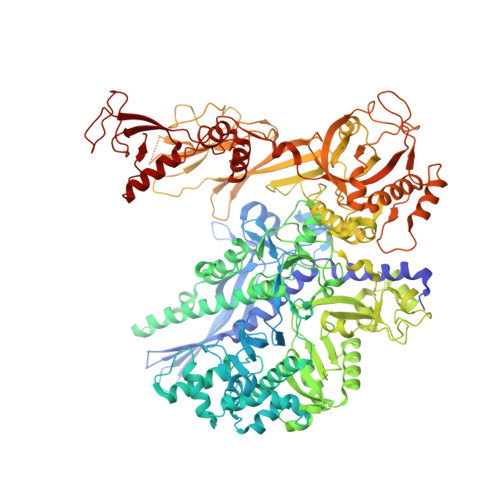
Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the structures of human TFIID-based PIC in three stepwise assembly states and revealed two-track PIC assembly: stepwise promoter deposition to Pol II and extensive modular reorganization on track I (on TATA-TFIID-binding element promoters) versus direct promoter deposition on track II (on TATA-only and TATA-less promoters). The two tracks converge at an ~50-subunit holo PIC in identical conformation, whereby TFIID stabilizes PIC organization and supports loading of cyclin-dependent kinase (CDK)-activating kinase (CAK) onto Pol II and CAK-mediated phosphorylation of the Pol II carboxyl-terminal domain. Unexpectedly, TBP of TFIID similarly bends TATA box and TATA-less promoters in PIC. Our study provides structural visualization of stepwise PIC assembly on highly diversified promoters.
- Fudan University Shanghai Cancer Center, Institutes of Biomedical Sciences, State Key Laboratory of Genetic Engineering and Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College of Fudan University, Shanghai 200032, China.
Organizational Affiliation: