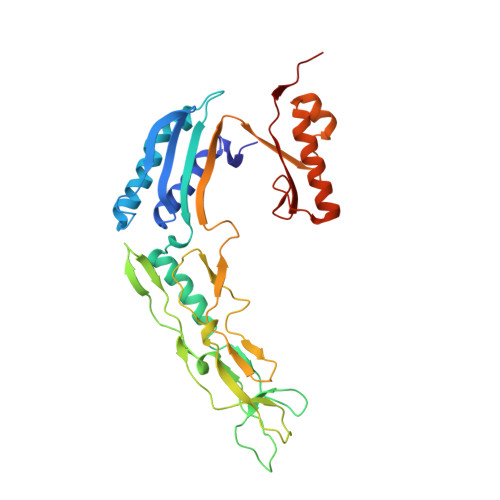
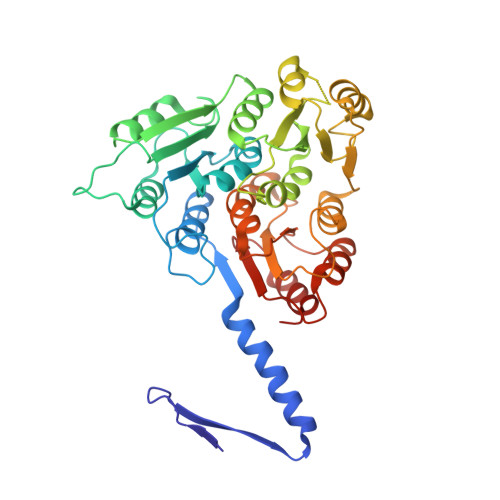
Structure and assembly of the S-layer in C. difficile.
Lanzoni-Mangutchi, P., Banerji, O., Wilson, J., Barwinska-Sendra, A., Kirk, J.A., Vaz, F., O'Beirne, S., Basle, A., El Omari, K., Wagner, A., Fairweather, N.F., Douce, G.R., Bullough, P.A., Fagan, R.P., Salgado, P.S.(2022) Nat Commun 13: 970-970
- PubMed: 35217634
- DOI: https://doi.org/10.1038/s41467-022-28196-w
- Primary Citation of Related Structures:
7ACV, 7ACW, 7ACX, 7ACY, 7ACZ, 7QGQ - PubMed Abstract:
Many bacteria and archaea possess a two-dimensional protein array, or S-layer, that covers the cell surface and plays crucial roles in cell physiology. Here, we report the crystal structure of SlpA, the main S-layer protein of the bacterial pathogen Clostridioides difficile, and use electron microscopy to study S-layer organisation and assembly. The SlpA crystal lattice mimics S-layer assembly in the cell, through tiling of triangular prisms above the cell wall, interlocked by distinct ridges facing the environment. Strikingly, the array is very compact, with pores of only ~10 Å in diameter, compared to other S-layers (30-100 Å). The surface-exposed flexible ridges are partially dispensable for overall structure and assembly, although a mutant lacking this region becomes susceptible to lysozyme, an important molecule in host defence. Thus, our work gives insights into S-layer organisation and provides a basis for development of C. difficile-specific therapeutics.
- Biosciences Institute, Faculty of Medical Sciences, Newcastle University, Newcastle upon Tyne, UK.
Organizational Affiliation: