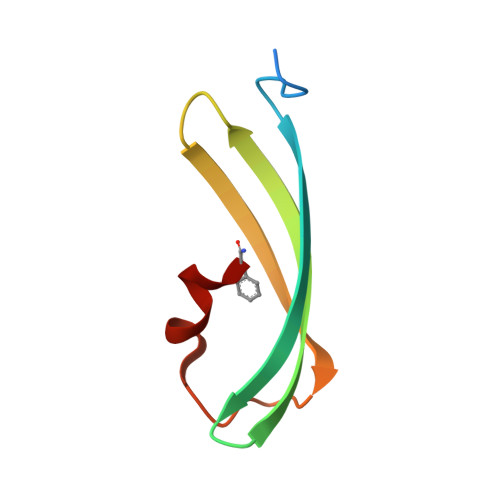
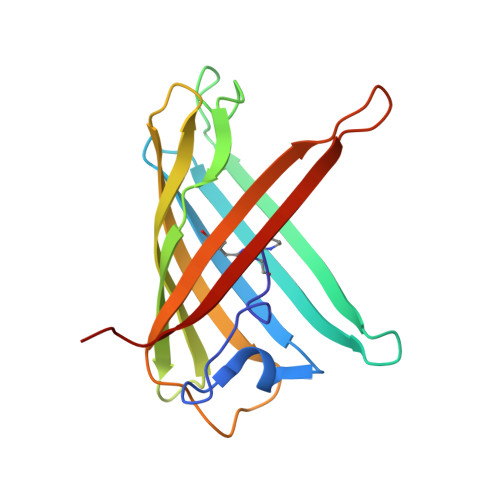
Spectroscopic and Structural Analysis of Cu 2+ -Induced Fluorescence Quenching of ZsYellow.
Kim, I.J., Xu, Y., Nam, K.H.(2020) Biosensors (Basel) 10
- PubMed: 32210006
- DOI: https://doi.org/10.3390/bios10030029
- Primary Citation of Related Structures:
6LOF - PubMed Abstract:
Fluorescent proteins exhibit fluorescence quenching by specific transition metals, suggesting their potential as fluorescent protein-based metal biosensors. Each fluorescent protein exhibits unique spectroscopic properties and mechanisms for fluorescence quenching by metals. Therefore, the metal-induced fluorescence quenching analysis of various new fluorescent proteins would be important step towards the development of such fluorescent protein-based metal biosensors. Here, we first report the spectroscopic and structural analysis of the yellow fluorescent protein ZsYellow, following its metal-induced quenching. Spectroscopic analysis showed that ZsYellow exhibited a high degree of fluorescence quenching by Cu 2+ . During Cu 2+ -induced ZsYellow quenching, fluorescence emission was recovered by adding EDTA. The crystal structure of ZsYellow soaked in Cu 2+ solution was determined at a 2.6 Å resolution. The electron density map did not indicate the presence of Cu 2+ around the chromophore or the β-barrel surface, which resulted in fluorescence quenching without Cu 2+ binding to specific site in ZsYellow. Based on these results, we propose the fluorescence quenching to occur in a distance-dependent manner between the metal and the fluorescent protein, when these components get to a closer vicinity at higher metal concentrations. Our results provide useful insights for future development of fluorescent protein-based metal biosensors.
- Division of Biotechnology, College of Life Sciences and Biotechnology, Korea University, Seoul 02841, Korea.
Organizational Affiliation: