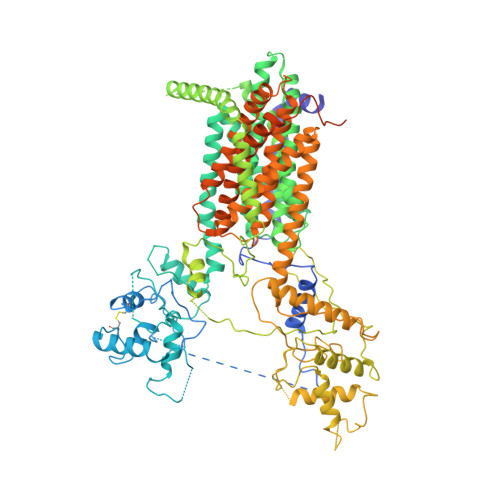
Structure of human Dispatched-1 provides insights into Hedgehog ligand biogenesis.
Chen, H., Liu, Y., Li, X.(2020) Life Sci Alliance 3
- PubMed: 32646883
- DOI: https://doi.org/10.26508/lsa.202000776
- Primary Citation of Related Structures:
6XE6 - PubMed Abstract:
Hedgehog (HH) signaling is essential for metazoan development. The HH ligand is secreted into the extracellular space by a cell surface protein named Dispatched-1 (DISP1). Here, we report the cryo-EM structure of human DISP1 protein. DISP1 contains 12 transmembrane helices (TMs) and two extracellular domains (ECDs). Its ECDs reveal an open state, in contrast to its structural homologues PTCH1 and NPC1, whose extracellular/luminal domains adopt a closed state. The low-resolution structure of the DISP1 complex with dual lipid-modified HH ligand reveals how the ECDs of DISP1 engage with HH ligand. Moreover, several cholesterol-like molecules are found in the TMs, implying a transport-like function of DISP1.
- Department of Molecular Genetics, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: