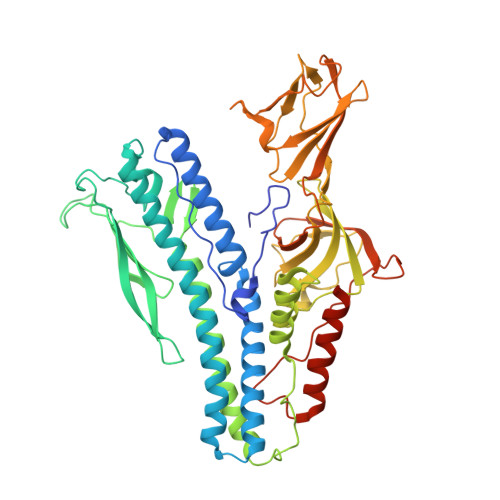
The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
El Omari, K., Li, S., Kotecha, A., Walter, T.S., Bignon, E.A., Harlos, K., Somerharju, P., De Haas, F., Clare, D.K., Molin, M., Hurtado, F., Li, M., Grimes, J.M., Bamford, D.H., Tischler, N.D., Huiskonen, J.T., Stuart, D.I., Roine, E.(2019) Nat Commun 10: 846-846
- PubMed: 30783086
- DOI: https://doi.org/10.1038/s41467-019-08728-7
- Primary Citation of Related Structures:
6J7V, 6QGI, 6QGL - PubMed Abstract:
Lipid membrane fusion is an essential function in many biological processes. Detailed mechanisms of membrane fusion and the protein structures involved have been mainly studied in eukaryotic systems, whereas very little is known about membrane fusion in prokaryotes. Haloarchaeal pleomorphic viruses (HRPVs) have a membrane envelope decorated with spikes that are presumed to be responsible for host attachment and membrane fusion. Here we determine atomic structures of the ectodomains of the 57-kDa spike protein VP5 from two related HRPVs revealing a previously unreported V-shaped fold. By Volta phase plate cryo-electron tomography we show that VP5 is monomeric on the viral surface, and we establish the orientation of the molecules with respect to the viral membrane. We also show that the viral membrane fuses with the host cytoplasmic membrane in a process mediated by VP5. This sheds light on protein structures involved in prokaryotic membrane fusion.
- Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford, OX3 7BN, UK.
Organizational Affiliation: