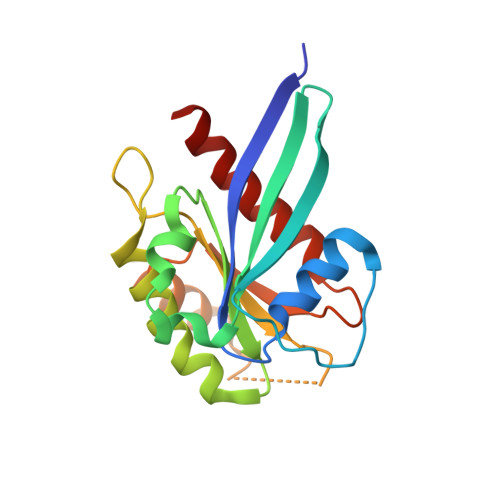
LRRK2 binds to the Rab32 subfamily in a GTP-dependent mannerviaits armadillo domain.
McGrath, E., Waschbusch, D., Baker, B.M., Khan, A.R.(2019) Small GTPases : 1-14
- PubMed: 31552791
- DOI: https://doi.org/10.1080/21541248.2019.1666623
- Primary Citation of Related Structures:
6FF8, 6HDU, 6HH2 - PubMed Abstract:
LRRK2 is a multi-domain Ser/Thr kinase that is associated with inherited and sporadic cases of Parkinson's disease. Many mutations linked to disease are associated within a central ROC-COR regulatory region and the subsequent kinase domain, leading to enhanced catalytic activity. The N-terminus of human LRRK2 consists of armadillo repeat motifs (ARMs) followed by ankyrin repeats (ANKs). Recently, Rab GTPases have emerged as key players in LRRK2 function, both as substrates of the kinase, and as regulators of the catalytic activity. Rabs recruit effector proteins via their GTP-dependent switch 1 and 2 regions to distinct sub-cellular compartments to regulate membrane trafficking. LRRK2 phosphorylates Rab8, Rab10 and Rab12 in switch 2, and this activity is regulated via interactions with Rab29. Furthermore, the related Rab32-subfamily GTPases, Rab32 and Rab38, have also been shown to interact with LRRK2. Here, we have mapped the interactions of the Rab32-subfamily to the ARM domain of LRRK2. The complexes are dependent on the GTP state of the Rabs in vitro , implying that LRRK2 may be an effector of the Rab32-subfamily of small GTPases. X-ray crystal structures of the Rab32-family GTPases and subsequent mutational studies reveal that a positively charged residue in switch 1 is critical for binding of Rab32/38 to LRRK2. Homology modelling and mutational analyses of the ARM domain point to a patch of negatively charged residues that contribute to complex formation. These structural and biochemical studies provide a framework for understanding the molecular basis for Rab regulation of LRRK2 and its role in Parkinson's disease.
- School of Biochemistry and Immunology, Trinity College Dublin , Dublin, Ireland.
Organizational Affiliation: