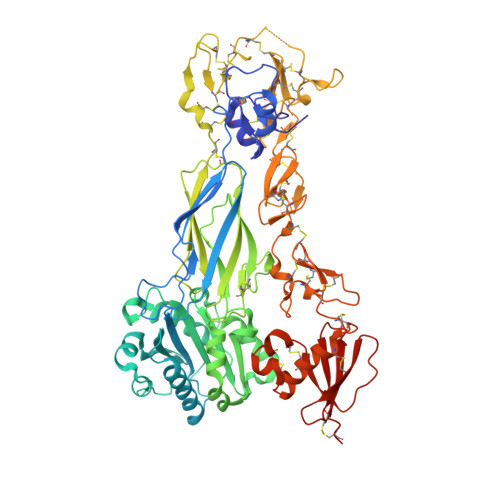
Autonomous conformational regulation of beta3integrin and the conformation-dependent property of HPA-1a alloantibodies.
Thinn, A.M.M., Wang, Z., Zhou, D., Zhao, Y., Curtis, B.R., Zhu, J.(2018) Proc Natl Acad Sci U S A 115: E9105-E9114
- PubMed: 30209215
- DOI: https://doi.org/10.1073/pnas.1806205115
- Primary Citation of Related Structures:
6BXJ - PubMed Abstract:
Integrin α/β heterodimer adopts a compact bent conformation in the resting state, and upon activation undergoes a large-scale conformational rearrangement. During the inside-out activation, signals impinging on the cytoplasmic tail of β subunit induce the α/β separation at the transmembrane and cytoplasmic domains, leading to the extended conformation of the ectodomain with the separated leg and the opening headpiece that is required for the high-affinity ligand binding. It remains enigmatic which integrin subunit drives the bent-to-extended conformational rearrangement in the inside-out activation. The β 3 integrins, including α IIb β 3 and α V β 3 , are the prototypes for understanding integrin structural regulation. The Leu33Pro polymorphism located at the β 3 PSI domain defines the human platelet-specific alloantigen (HPA) 1a/b, which provokes the alloimmune response leading to clinically important bleeding disorders. Some, but not all, anti-HPA-1a alloantibodies can distinguish the α IIb β 3 from α V β 3 and affect their functions with unknown mechanisms. Here we designed a single-chain β 3 subunit that mimics a separation of α/β heterodimer on inside-out activation. Our crystallographic and functional studies show that the single-chain β 3 integrin folds into a bent conformation in solution but spontaneously extends on the cell surface. This demonstrates that the β 3 subunit autonomously drives the membrane-dependent conformational rearrangement during integrin activation. Using the single-chain β 3 integrin, we identified the conformation-dependent property of anti-HPA-1a alloantibodies, which enables them to differently recognize the β 3 in the bent state vs. the extended state and in the complex with α IIb vs. α V This study provides deeper understandings of integrin conformational activation on the cell surface.
- Blood Research Institute, BloodCenter of Wisconsin, Part of Versiti, Milwaukee, WI 53226.
Organizational Affiliation: