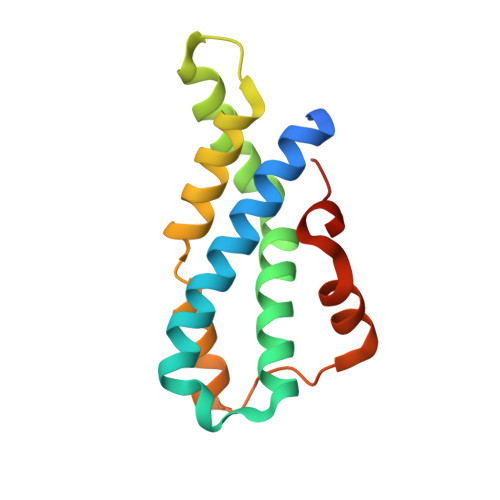
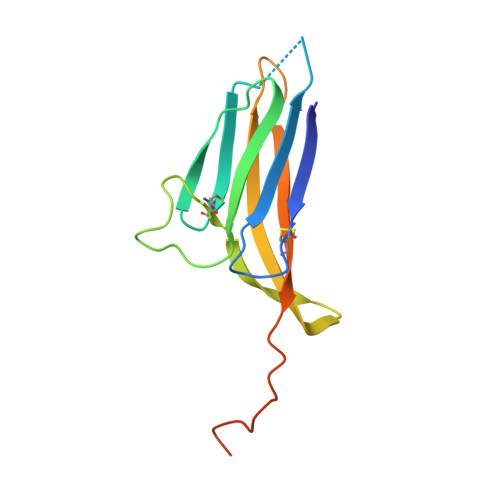
Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Raj, I., Sadat Al Hosseini, H., Dioguardi, E., Nishimura, K., Han, L., Villa, A., de Sanctis, D., Jovine, L.(2017) Cell 169: 1315-1326.e17
- PubMed: 28622512
- DOI: https://doi.org/10.1016/j.cell.2017.05.033
- Primary Citation of Related Structures:
5II4, 5II5, 5II6, 5II7, 5II8, 5II9, 5IIA, 5IIB, 5IIC, 5MR2, 5MR3 - PubMed Abstract:
Recognition between sperm and the egg surface marks the beginning of life in all sexually reproducing organisms. This fundamental biological event depends on the species-specific interaction between rapidly evolving counterpart molecules on the gametes. We report biochemical, crystallographic, and mutational studies of domain repeats 1-3 of invertebrate egg coat protein VERL and their interaction with cognate sperm protein lysin. VERL repeats fold like the functionally essential N-terminal repeat of mammalian sperm receptor ZP2, whose structure is also described here. Whereas sequence-divergent repeat 1 does not bind lysin, repeat 3 binds it non-species specifically via a high-affinity, largely hydrophobic interface. Due to its intermediate binding affinity, repeat 2 selectively interacts with lysin from the same species. Exposure of a highly positively charged surface of VERL-bound lysin suggests that complex formation both disrupts the organization of egg coat filaments and triggers their electrostatic repulsion, thereby opening a hole for sperm penetration and fusion.
- Department of Biosciences and Nutrition and Center for Innovative Medicine, Karolinska Institutet, Huddinge, SE-141 83, Sweden.
Organizational Affiliation: