Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Johannes, J.W., Bates, S., Beigie, C., Belmonte, M., Breen, J., Cao, S., Centrella, P.A., Clark, M.A., Cuozzo, J.W., Dumelin, C.E., Ferguson, A.D., Habeshian, S., Hargreaves, D., Joubran, C., Kazmirski, S., Keefe, A., Lamb, M.L., Lan, H., Li, Y., Ma, H., Mlynarski, S., Packer, M.J., Rawlins, P., Robbins, D.W., Shen, H., Sigel, E.A., Soutter, H.H., Su, N., Trost, D.M., Wang, H., Wickson, K.F., Wu, C., Zhang, Y., Zhao, Q., Zheng, X., Hird, A.(2017) ACS Med Chem Lett 8: 239-244
- PubMed: 28197319
- DOI: https://doi.org/10.1021/acsmedchemlett.6b00464
- Primary Citation of Related Structures:
5KU9, 5MES, 5MEV - PubMed Abstract:
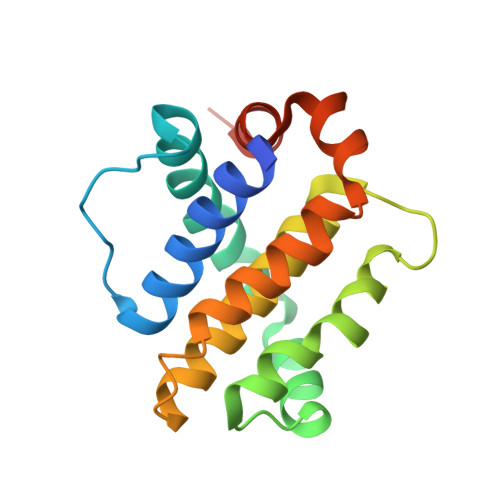
Mcl-1 is a pro-apoptotic BH3 protein family member similar to Bcl-2 and Bcl-xL. Overexpression of Mcl-1 is often seen in various tumors and allows cancer cells to evade apoptosis. Here we report the discovery and optimization of a series of non-natural peptide Mcl-1 inhibitors. Screening of DNA-encoded libraries resulted in hit compound 1 , a 1.5 μM Mcl-1 inhibitor. A subsequent crystal structure demonstrated that compound 1 bound to Mcl-1 in a β-turn conformation, such that the two ends of the peptide were close together. This proximity allowed for the linking of the two ends of the peptide to form a macrocycle. Macrocyclization resulted in an approximately 10-fold improvement in binding potency. Further exploration of a key hydrophobic interaction with Mcl-1 protein and also with the moiety that engages Arg256 led to additional potency improvements. The use of protein-ligand crystal structures and binding kinetics contributed to the design and understanding of the potency gains. Optimized compound 26 is a <3 nM Mcl-1 inhibitor, while inhibiting Bcl-2 at only 5 μM and Bcl-xL at >99 μM, and induces cleaved caspase-3 in MV4-11 cells with an IC 50 of 3 μM after 6 h.
- AstraZeneca R&D Boston , 35 Gatehouse Drive, Waltham, Massachusetts 02451, United States.
Organizational Affiliation: