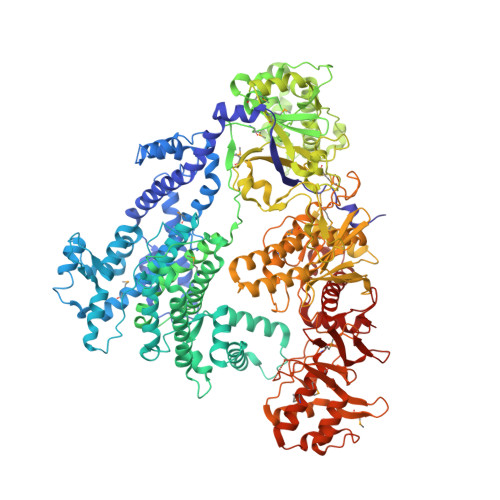
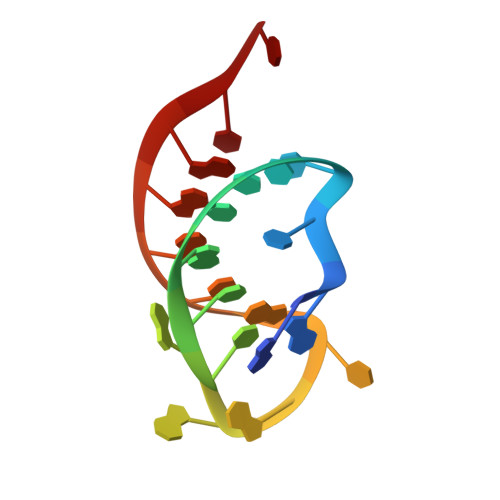
The crystal structure of Cpf1 in complex with CRISPR RNA
Dong, D., Ren, K., Qiu, X., Zheng, J., Guo, M., Guan, X., Liu, H., Li, N., Zhang, B., Yang, D., Ma, C., Wang, S., Wu, D., Ma, Y., Fan, S., Wang, J., Gao, N., Huang, Z.(2016) Nature 532: 522-526
- PubMed: 27096363
- DOI: https://doi.org/10.1038/nature17944
- Primary Citation of Related Structures:
5ID6 - PubMed Abstract:
The CRISPR-Cas systems, as exemplified by CRISPR-Cas9, are RNA-guided adaptive immune systems used by bacteria and archaea to defend against viral infection. The CRISPR-Cpf1 system, a new class 2 CRISPR-Cas system, mediates robust DNA interference in human cells. Although functionally conserved, Cpf1 and Cas9 differ in many aspects including their guide RNAs and substrate specificity. Here we report the 2.38 Å crystal structure of the CRISPR RNA (crRNA)-bound Lachnospiraceae bacterium ND2006 Cpf1 (LbCpf1). LbCpf1 has a triangle-shaped architecture with a large positively charged channel at the centre. Recognized by the oligonucleotide-binding domain of LbCpf1, the crRNA adopts a highly distorted conformation stabilized by extensive intramolecular interactions and the (Mg(H2O)6)(2+) ion. The oligonucleotide-binding domain also harbours a looped-out helical domain that is important for LbCpf1 substrate binding. Binding of crRNA or crRNA lacking the guide sequence induces marked conformational changes but no oligomerization of LbCpf1. Our study reveals the crRNA recognition mechanism and provides insight into crRNA-guided substrate binding of LbCpf1, establishing a framework for engineering LbCpf1 to improve its efficiency and specificity for genome editing.
- School of Life Science and Technology, Harbin Institute of Technology, Harbin 150080, China.
Organizational Affiliation: