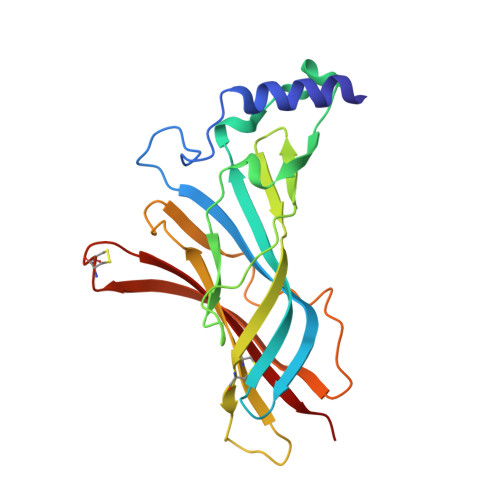
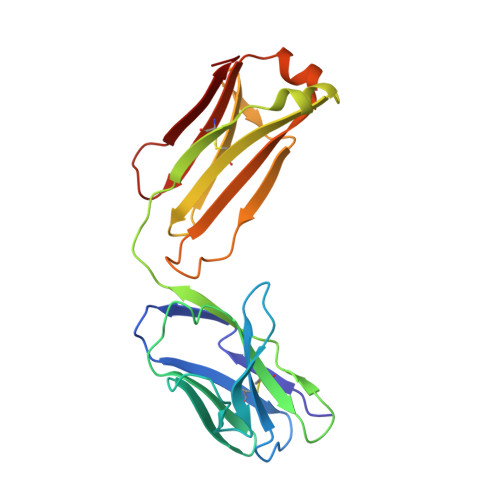
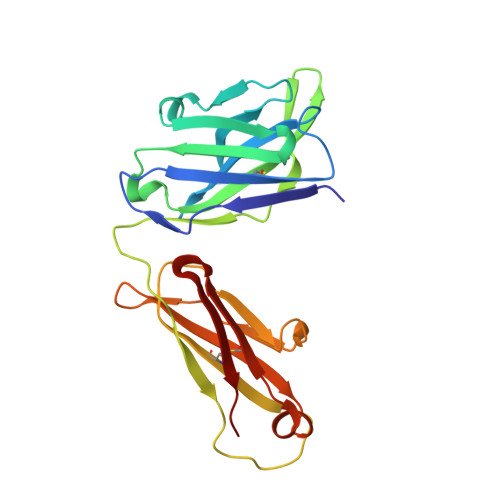
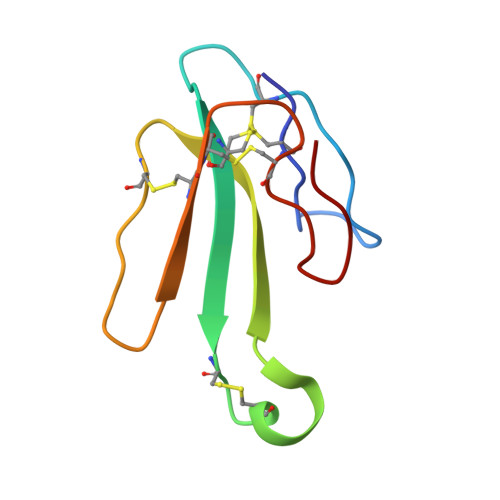
Structural insights into the molecular mechanisms of myasthenia gravis and their therapeutic implications.
Noridomi, K., Watanabe, G., Hansen, M.N., Han, G.W., Chen, L.(2017) Elife 6
- PubMed: 28440223
- DOI: https://doi.org/10.7554/eLife.23043
- Primary Citation of Related Structures:
5HBT, 5HBV - PubMed Abstract:
The nicotinic acetylcholine receptor (nAChR) is a major target of autoantibodies in myasthenia gravis (MG), an autoimmune disease that causes neuromuscular transmission dysfunction. Despite decades of research, the molecular mechanisms underlying MG have not been fully elucidated. Here, we present the crystal structure of the nAChR α1 subunit bound by the Fab fragment of mAb35, a reference monoclonal antibody that causes experimental MG and competes with ~65% of antibodies from MG patients. Our structures reveal for the first time the detailed molecular interactions between MG antibodies and a core region on nAChR α1. These structures suggest a major nAChR-binding mechanism shared by a large number of MG antibodies and the possibility to treat MG by blocking this binding mechanism. Structure-based modeling also provides insights into antibody-mediated nAChR cross-linking known to cause receptor degradation. Our studies establish a structural basis for further mechanistic studies and therapeutic development of MG.
- Department of Chemistry, University of Southern California, Los Angeles, United States.
Organizational Affiliation: