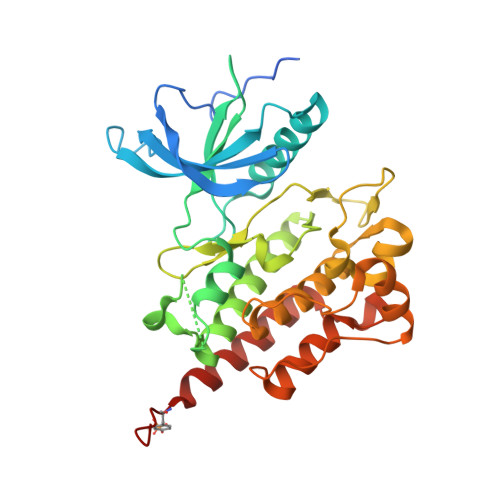
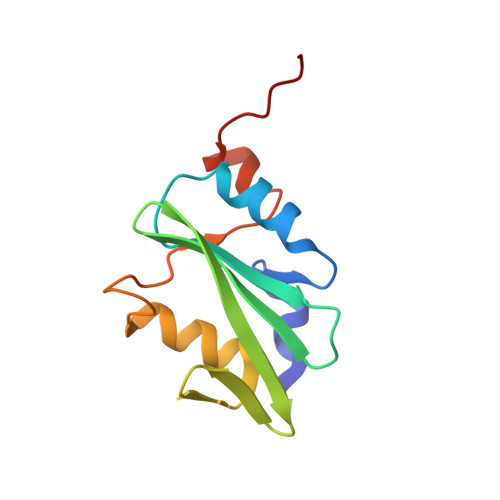
Two FGF Receptor Kinase Molecules Act in Concert to Recruit and Transphosphorylate Phospholipase C gamma.
Huang, Z., Marsiglia, W.M., Basu Roy, U., Rahimi, N., Ilghari, D., Wang, H., Chen, H., Gai, W., Blais, S., Neubert, T.A., Mansukhani, A., Traaseth, N.J., Li, X., Mohammadi, M.(2016) Mol Cell 61: 98-110
- PubMed: 26687682
- DOI: https://doi.org/10.1016/j.molcel.2015.11.010
- Primary Citation of Related Structures:
5EG3 - PubMed Abstract:
The molecular basis by which receptor tyrosine kinases (RTKs) recruit and phosphorylate Src Homology 2 (SH2) domain-containing substrates has remained elusive. We used X-ray crystallography, NMR spectroscopy, and cell-based assays to demonstrate that recruitment and phosphorylation of Phospholipase Cγ (PLCγ), a prototypical SH2 containing substrate, by FGF receptors (FGFR) entails formation of an allosteric 2:1 FGFR-PLCγ complex. We show that the engagement of pTyr-binding pocket of the cSH2 domain of PLCγ by the phosphorylated tail of an FGFR kinase induces a conformational change at the region past the cSH2 core domain encompassing Tyr-771 and Tyr-783 to facilitate the binding/phosphorylation of these tyrosines by another FGFR kinase in trans. Our data overturn the current paradigm that recruitment and phosphorylation of substrates are carried out by the same RTK monomer in cis and disclose an obligatory role for receptor dimerization in substrate phosphorylation in addition to its canonical role in kinase activation.
- School of Pharmacy, Wenzhou Medical University, Wenzhou, Zhejiang 325035, China; Department of Biochemistry & Molecular Pharmacology, New York University School of Medicine, New York, NY 10016, USA.
Organizational Affiliation: