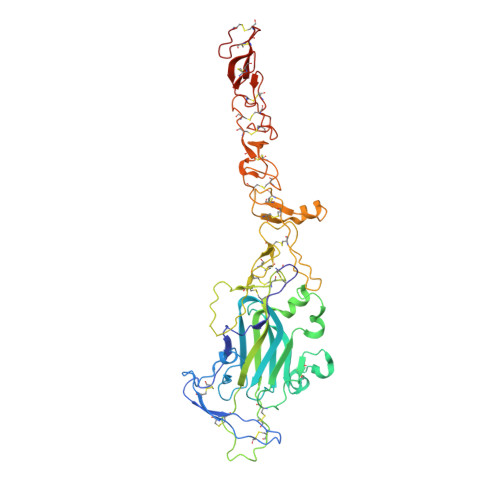
Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes.
Reuten, R., Patel, T.R., McDougall, M., Rama, N., Nikodemus, D., Gibert, B., Delcros, J.G., Prein, C., Meier, M., Metzger, S., Zhou, Z., Kaltenberg, J., McKee, K.K., Bald, T., Tuting, T., Zigrino, P., Djonov, V., Bloch, W., Clausen-Schaumann, H., Poschl, E., Yurchenco, P.D., Ehrbar, M., Mehlen, P., Stetefeld, J., Koch, M.(2016) Nat Commun 7: 13515-13515
- PubMed: 27901020
- DOI: https://doi.org/10.1038/ncomms13515
- Primary Citation of Related Structures:
4WNX - PubMed Abstract:
Netrins, a family of laminin-related molecules, have been proposed to act as guidance cues either during nervous system development or the establishment of the vascular system. This was clearly demonstrated for netrin-1 via its interaction with the receptors DCC and UNC5s. However, mainly based on shared homologies with netrin-1, netrin-4 was also proposed to play a role in neuronal outgrowth and developmental/pathological angiogenesis via interactions with netrin-1 receptors. Here, we present the high-resolution structure of netrin-4, which shows unique features in comparison with netrin-1, and show that it does not bind directly to any of the known netrin-1 receptors. We show that netrin-4 disrupts laminin networks and basement membranes (BMs) through high-affinity binding to the laminin γ1 chain. We hypothesize that this laminin-related function is essential for the previously described effects on axon growth promotion and angiogenesis. Our study unveils netrin-4 as a non-enzymatic extracellular matrix protein actively disrupting pre-existing BMs.
- Institute for Dental Research and Oral Musculoskeletal Biology, Medical Faculty, University of Cologne, Joseph-Stelzmann-Strasse 52, Cologne 50931, Germany.
Organizational Affiliation: