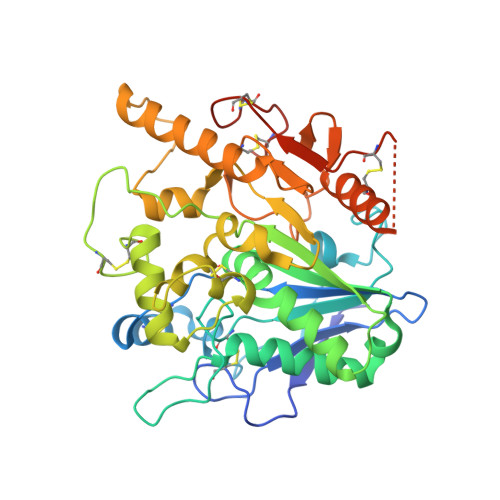
Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Kakugawa, S., Langton, P.F., Zebisch, M., Howell, S.A., Chang, T., Liu, Y., Feizi, T., Bineva, G., O'Reilly, N., Snijders, A.P., Jones, E.Y., Vincent, J.(2015) Nature 519: 187
- PubMed: 25731175
- DOI: https://doi.org/10.1038/nature14259
- Primary Citation of Related Structures:
4UYU, 4UYW, 4UYZ, 4UZ1, 4UZ5, 4UZ6, 4UZ7, 4UZ9, 4UZA, 4UZJ, 4UZK, 4UZL, 4UZQ, 4WBH - PubMed Abstract:
Signalling by Wnt proteins is finely balanced to ensure normal development and tissue homeostasis while avoiding diseases such as cancer. This is achieved in part by Notum, a highly conserved secreted feedback antagonist. Notum has been thought to act as a phospholipase, shedding glypicans and associated Wnt proteins from the cell surface. However, this view fails to explain specificity, as glypicans bind many extracellular ligands. Here we provide genetic evidence in Drosophila that Notum requires glypicans to suppress Wnt signalling, but does not cleave their glycophosphatidylinositol anchor. Structural analyses reveal glycosaminoglycan binding sites on Notum, which probably help Notum to co-localize with Wnt proteins. They also identify, at the active site of human and Drosophila Notum, a large hydrophobic pocket that accommodates palmitoleate. Kinetic and mass spectrometric analyses of human proteins show that Notum is a carboxylesterase that removes an essential palmitoleate moiety from Wnt proteins and thus constitutes the first known extracellular protein deacylase.
- MRC's National Institute for Medical Research, The Ridgeway, Mill Hill, London NW7 1AA, UK.
Organizational Affiliation: