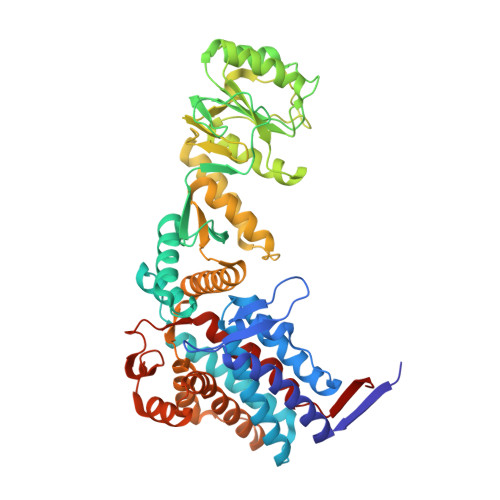
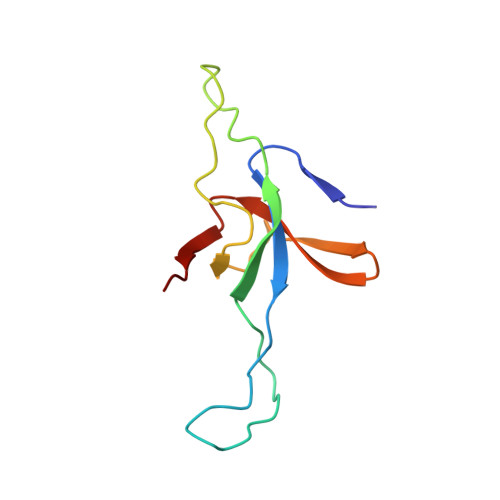
Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Chen, D.-H., Madan, D., Weaver, J., Lin, Z., Schroder, G.F., Chiu, W., Rye, H.S.(2013) Cell 153: 1354
- PubMed: 23746846
- DOI: https://doi.org/10.1016/j.cell.2013.04.052
- Primary Citation of Related Structures:
3ZPZ, 3ZQ0, 3ZQ1 - PubMed Abstract:
The GroEL/ES chaperonin system is required for the assisted folding of many proteins. How these substrate proteins are encapsulated within the GroEL-GroES cavity is poorly understood. Using symmetry-free, single-particle cryo-electron microscopy, we have characterized a chemically modified mutant of GroEL (EL43Py) that is trapped at a normally transient stage of substrate protein encapsulation. We show that the symmetric pattern of the GroEL subunits is broken as the GroEL cis-ring apical domains reorient to accommodate the simultaneous binding of GroES and an incompletely folded substrate protein (RuBisCO). The collapsed RuBisCO folding intermediate binds to the lower segment of two apical domains, as well as to the normally unstructured GroEL C-terminal tails. A comparative structural analysis suggests that the allosteric transitions leading to substrate protein release and folding involve concerted shifts of GroES and the GroEL apical domains and C-terminal tails.
- Verna and Marrs McLean Department of Biochemistry and Molecular Biology, National Center for Macromolecular Imaging, Baylor College of Medicine, Houston, TX 77030, USA.
Organizational Affiliation: