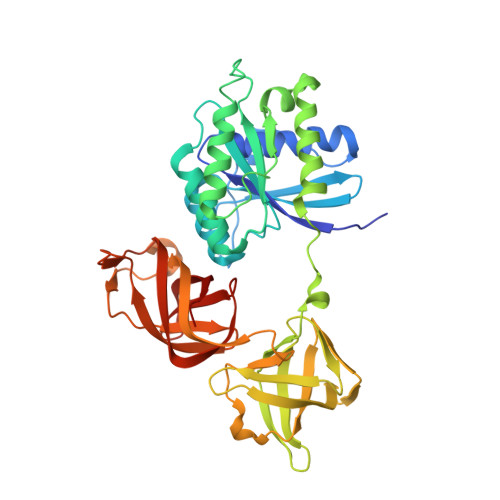
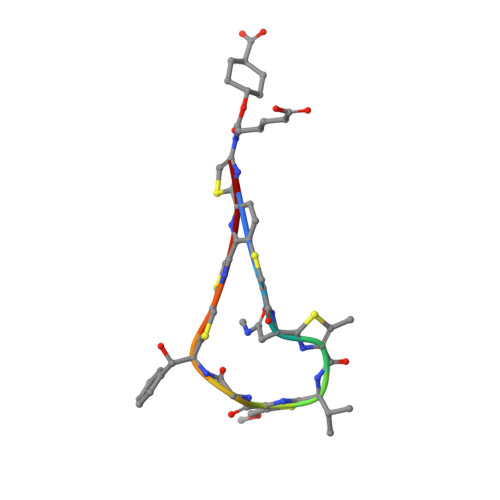
Discovery of LFF571: an investigational agent for Clostridium difficile infection.
LaMarche, M.J., Leeds, J.A., Amaral, A., Brewer, J.T., Bushell, S.M., Deng, G., Dewhurst, J.M., Ding, J., Dzink-Fox, J., Gamber, G., Jain, A., Lee, K., Lee, L., Lister, T., McKenney, D., Mullin, S., Osborne, C., Palestrant, D., Patane, M.A., Rann, E.M., Sachdeva, M., Shao, J., Tiamfook, S., Trzasko, A., Whitehead, L., Yifru, A., Yu, D., Yan, W., Zhu, Q.(2012) J Med Chem 55: 2376-2387
- PubMed: 22315981
- DOI: https://doi.org/10.1021/jm201685h
- Primary Citation of Related Structures:
3U2Q - PubMed Abstract:
Clostridium difficile (C. difficile) is a Gram positive, anaerobic bacterium that infects the lumen of the large intestine and produces toxins. This results in a range of syndromes from mild diarrhea to severe toxic megacolon and death. Alarmingly, the prevalence and severity of C. difficile infection are increasing; thus, associated morbidity and mortality rates are rising. 4-Aminothiazolyl analogues of the antibiotic natural product GE2270 A (1) were designed, synthesized, and optimized for the treatment of C. difficile infection. The medicinal chemistry effort focused on enhancing aqueous solubility relative to that of the natural product and previous development candidates (2, 3) and improving antibacterial activity. Structure-activity relationships, cocrystallographic interactions, pharmacokinetics, and efficacy in animal models of infection were characterized. These studies identified a series of dicarboxylic acid derivatives, which enhanced solubility/efficacy profile by several orders of magnitude compared to previously studied compounds and led to the selection of LFF571 (4) as an investigational new drug for treating C. difficile infection.
- Global Discovery Chemistry, Novartis Institutes for Biomedical Research, 250 Massachusetts Avenue, Cambridge, Massachusetts 02139, USA. matthew.lamarche@novartis.com
Organizational Affiliation: