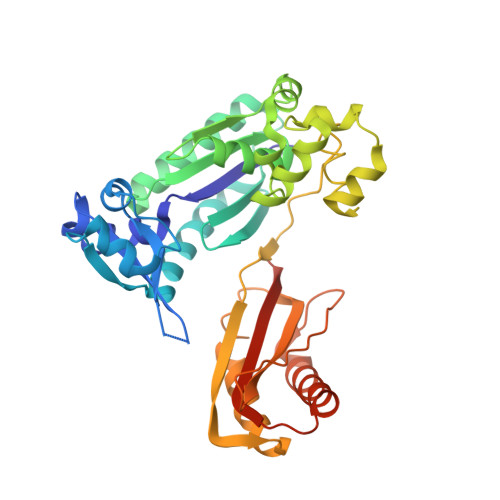
Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh.
Wilson, R.C., Pata, J.D.(2008) Mol Cell 29: 767-779
- PubMed: 18374650
- DOI: https://doi.org/10.1016/j.molcel.2008.01.014
- Primary Citation of Related Structures:
3BQ0, 3BQ1, 3BQ2 - PubMed Abstract:
Dbh is a Y family translesion DNA polymerase that accurately bypasses some damaged forms of deoxyguanosine, but also generates single-base deletion errors at frequencies of up to 50%, in specific hot spot sequences. We describe preinsertion binary, insertion ternary, and postinsertion binary crystal structures of Dbh synthesizing DNA after making a single-base deletion. The skipped template base adopts an extrahelical conformation stabilized by interactions with the C-terminal domain of the enzyme. DNA translocation and positioning of the next templating base at the active site, with space opposite to accommodate incoming nucleotide, occur independently of nucleotide binding, incorporation, and pyrophosphate release. We also show that Dbh creates single-base deletions more rapidly when the skipped base is located two or three bases upstream of the nascent base pair than when it is directly adjacent to the templating base, indicating that Dbh predominantly creates single-base deletions by template slippage rather than by dNTP-stabilized misalignment.
- Division of Molecular Medicine, Wadsworth Center, New York State Department of Health, The State University of New York at Albany, Albany, NY 12201-0509, USA.
Organizational Affiliation: