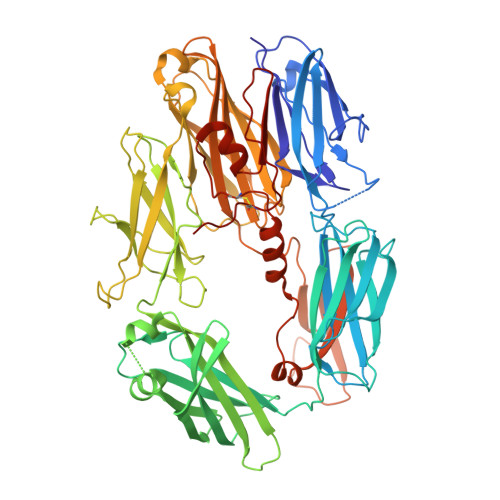
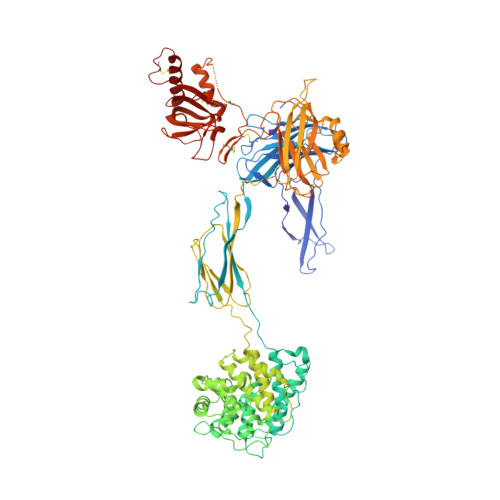
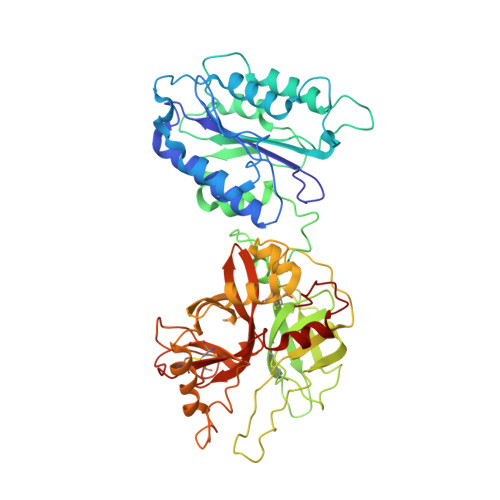
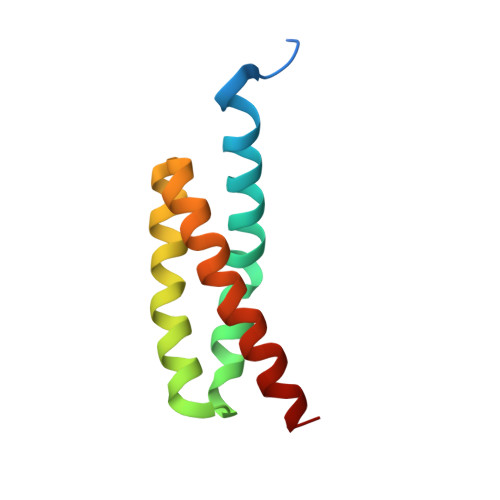
Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor.
Rooijakkers, S.H., Wu, J., Ruyken, M., van Domselaar, R., Planken, K.L., Tzekou, A., Ricklin, D., Lambris, J.D., Janssen, B.J., van Strijp, J.A., Gros, P.(2009) Nat Immunol 10: 721-727
- PubMed: 19503103
- DOI: https://doi.org/10.1038/ni.1756
- Primary Citation of Related Structures:
2WIN - PubMed Abstract:
Activation of the complement system generates potent chemoattractants and leads to the opsonization of cells for immune clearance. Short-lived protease complexes cleave complement component C3 into anaphylatoxin C3a and opsonin C3b. Here we report the crystal structure of the C3 convertase formed by C3b and the protease fragment Bb, which was stabilized by the bacterial immune-evasion protein SCIN. The data suggest that the proteolytic specificity and activity depend on the formation of dimers of C3 with C3b of the convertase. SCIN blocked the formation of a productive enzyme-substrate complex. Irreversible dissociation of the complex of C3b and Bb is crucial to complement regulation and was determined by slow binding kinetics of the Mg(2+)-adhesion site in Bb. Understanding the mechanistic basis of the central complement-activation step and microbial immune evasion strategies targeting this step will aid in the development of complement therapeutics.
- Medical Microbiology, University Medical Center Utrecht, Utrecht, The Netherlands.
Organizational Affiliation: