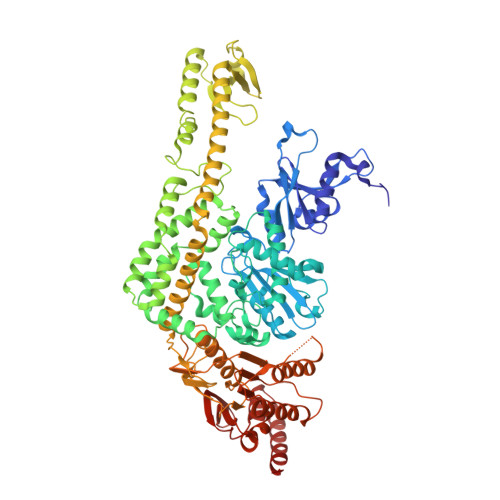
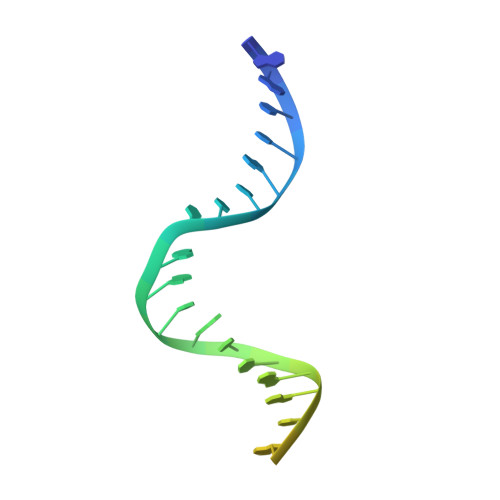
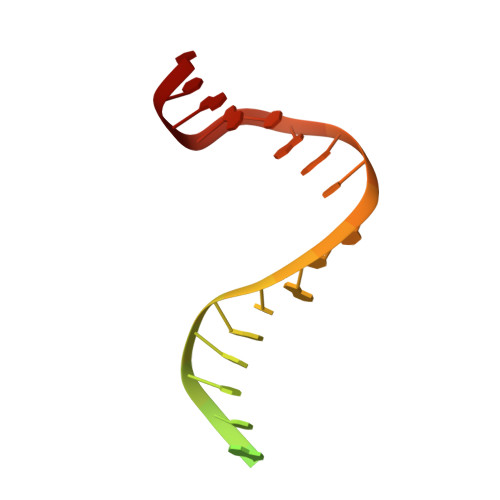
The alternating ATPase domains of MutS control DNA mismatch repair
Lamers, M.H., Winterwerp, H.H.K., Sixma, T.K.(2003) EMBO J 22: 746-756
- PubMed: 12554674
- DOI: https://doi.org/10.1093/emboj/cdg064
- Primary Citation of Related Structures:
1NG9 - PubMed Abstract:
DNA mismatch repair is an essential safeguard of genomic integrity by removing base mispairings that may arise from DNA polymerase errors or from homologous recombination between DNA strands. In Escherichia coli, the MutS enzyme recognizes mismatches and initiates repair. MutS has an intrinsic ATPase activity crucial for its function, but which is poorly understood. We show here that within the MutS homodimer, the two chemically identical ATPase sites have different affinities for ADP, and the two sites alternate in ATP hydrolysis. A single residue, Arg697, located at the interface of the two ATPase domains, controls the asymmetry. When mutated, the asymmetry is lost and mismatch repair in vivo is impaired. We propose that asymmetry of the ATPase domains is an essential feature of mismatch repair that controls the timing of the different steps in the repair cascade.
- Division of Molecular Carcinogenesis, Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Organizational Affiliation: