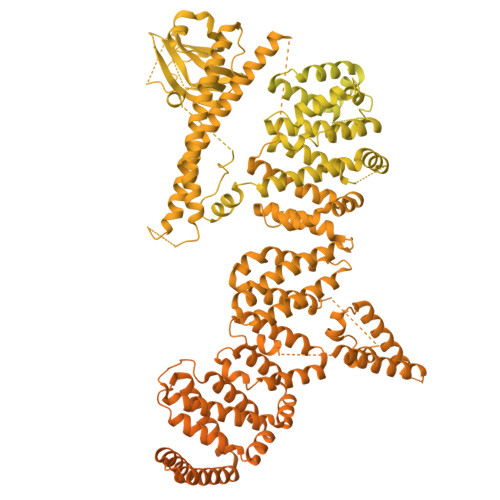
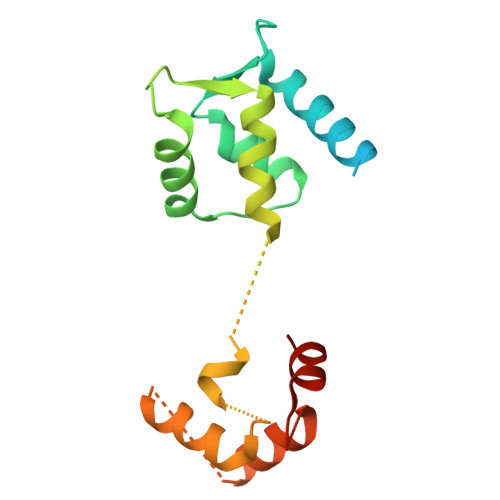
Architecture of the UBR4 complex, a giant E4 ligase central to eukaryotic protein quality control.
Grabarczyk, D.B., Ehrmann, J.F., Murphy, P., Yang, W.S., Kurzbauer, R., Bell, L.E., Deszcz, L., Neuhold, J., Schleiffer, A., Shulkina, A., Lee, J., Shin, J.S., Meinhart, A., Versteeg, G.A., Zavodszky, E., Song, H.K., Hegde, R.S., Clausen, T.(2025) Science 389: 909-914
- PubMed: 40875847
- DOI: https://doi.org/10.1126/science.adv9309
- Primary Citation of Related Structures:
9JNI, 9LGS, 9QT9, 9QWS, 9QWU, 9QWX, 9QWZ, 9QX0, 9QX1, 9QX2, 9QX5, 9UPZ - PubMed Abstract:
Eukaryotic cells have evolved sophisticated quality control mechanisms to eliminate aggregation-prone proteins that compromise cellular health. Central to this defense is the ubiquitin-proteasome system, where UBR4 acts as an essential E4 ubiquitin ligase, amplifying degradation marks on defective proteins. Cryo-electron microscopy analysis of UBR4 in complex with its cofactors KCMF1 and CALM1 reveals a massive 1.3-megadalton ring structure, featuring a central substrate-binding arena and flexibly attached catalytic units. Our structure shows how UBR4 binds substrate and extends lysine-48-specific ubiquitin chains. Efficient substrate targeting depends on both preubiquitination and specific N-degrons, with KCMF1 acting as a key substrate filter. The architecture of the E4 megacomplex is conserved across eukaryotes, but species-specific adaptations allow UBR4 to perform its precisely tuned quality control function in diverse cellular environments.
- Research Institute of Molecular Pathology, Vienna BioCenter (VBC), Vienna, Austria.
Organizational Affiliation: