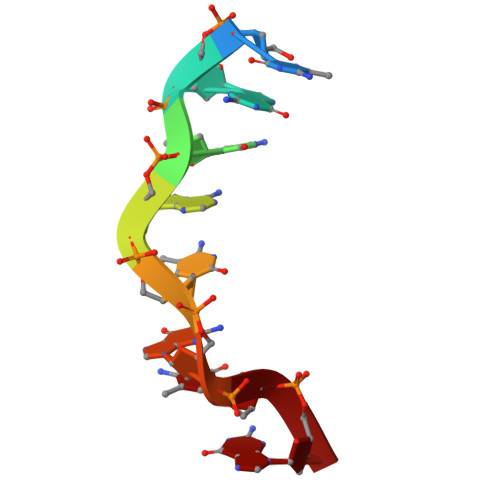
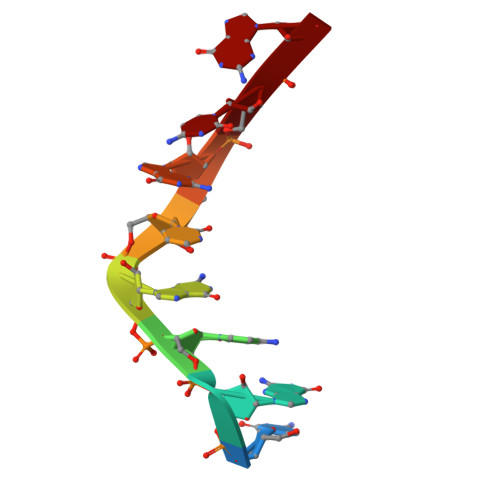
Z-form DNA-RNA hybrid blocks DNA replication.
Wang, S., Xu, Y.(2025) Nucleic Acids Res 53
- PubMed: 40037715
- DOI: https://doi.org/10.1093/nar/gkaf135
- Primary Citation of Related Structures:
9JVN, 9KZP - PubMed Abstract:
We discovered that the Z-form DNA-RNA hybrid stabilized by methylated CpG repeats impacts on the initiation and elongation of Okazaki fragments, contributing to blocking DNA replication at first time. We further present the first Z-form DNA-RNA hybrid structure by using NMR spectroscopy and dynamic computation, revealing the molecular mechanism of inhibition, indicating that a distinctive zig-zag strand pattern of the Z-form hybrid with a smaller helical diameter (15 Å) and a very narrow minor groove (8.3 Å) plays the key role in the repression toward DNA replication.
- Division of Chemistry, Department of Medical Sciences, Faculty of Medicine, University of Miyazaki, 5200 Kihara, Kiyotake, Miyazaki 889-1692, Japan.
Organizational Affiliation: