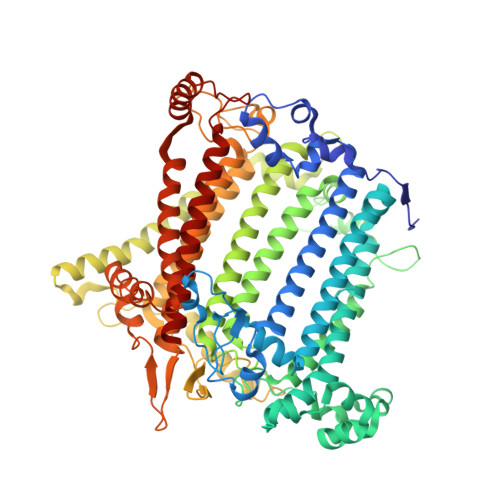
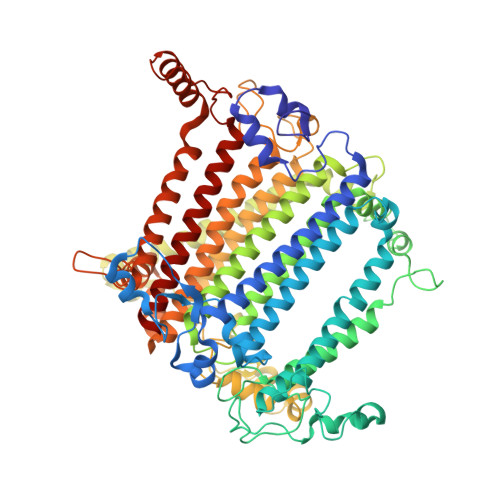
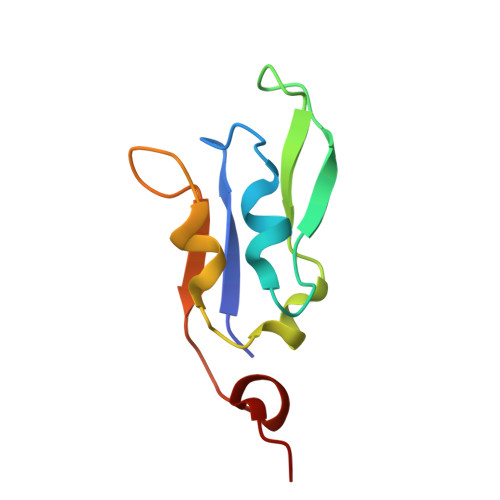
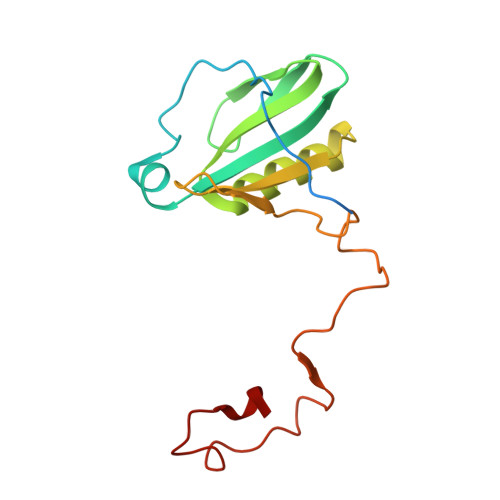
Structural determinants for red-shifted absorption in higher-plants Photosystem I.
Capaldi, S., Guardini, Z., Montepietra, D., Pagliuca, V.F., Amelii, A., Betti, E., John, C., Pedraza-Gonzalez, L., Cupellini, L., Mennucci, B., Bonnet, D.M.V., Chaves-Sanjuan, A., Dall'Osto, L., Bassi, R.(2025) New Phytol 248: 2331-2346
- PubMed: 40955088
- DOI: https://doi.org/10.1111/nph.70562
- Primary Citation of Related Structures:
9GBI, 9GC2 - PubMed Abstract:
Higher plants Photosystem I absorbs far-red light, enriched under vegetation canopies, through long-wavelength Chls to enhance photon capture. Far-red absorption originates from Chl pairs within the Lhca3 and Lhca4 subunits of the LHCI antenna, known as the 'red cluster', including Chls a603 and a609. We used reverse genetics to produce an Arabidopsis mutant devoid of red-shifted absorption, and we obtained high-resolution cryogenic electron microscopy structures of PSI-LHCI complexes from both wild-type and mutant plants. Computed excitonic coupling values suggested contributions from additional nearby pigment molecules, namely Chl a615 and violaxanthin in the L2 site, to far-red absorption. We investigated the structural determinants of far-red absorption by producing further Arabidopsis transgenic lines and analyzed the spectroscopic effects of mutations targeting these chromophores. The two structures solved were used for quantum mechanics calculations, revealing that excitonic interactions alone cannot explain far-red absorption, while charge transfer states were needed for accurate spectral simulations. Our findings demonstrate that the molecular mechanisms of light-harvesting under shaded conditions rely on very precise tuning of chromophore interactions, whose understanding is crucial for designing light-harvesting complexes with engineered absorption spectra.
- Dipartimento di Biotecnologie, Università di Verona, Strada Le Grazie 15, 37134, Verona, Italy.
Organizational Affiliation: