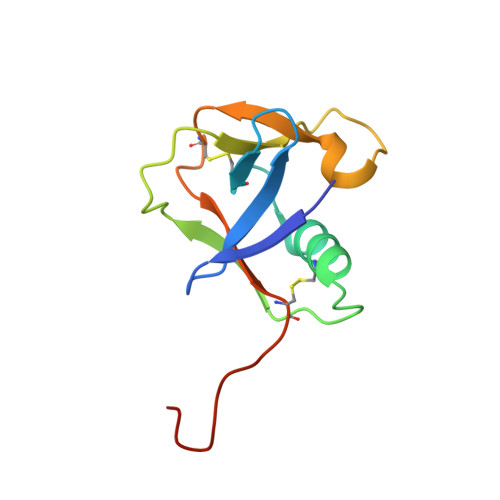
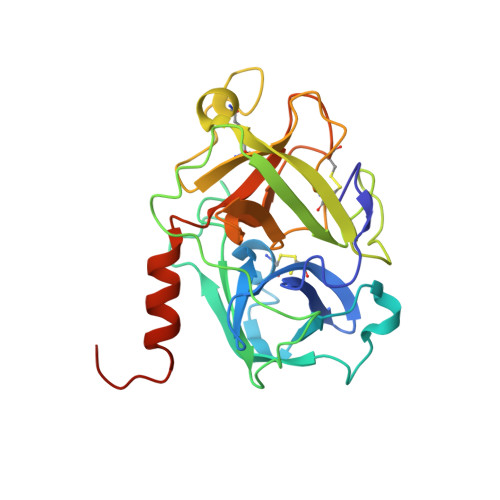
Large Library Docking and Biophysical Analysis of Small-Molecule TMPRSS2 Inhibitors.
Fraser, B.J., Young, N.J., Bender, B.J., Gahbauer, S., Ilyassov, O., Wilson, R.P., Li, Y., Seitova, A., Lourenco, A.L., Chung, D.H., Bardine, C., Benard, F., Shoichet, B.K., Craik, C.S., Arrowsmith, C.H.(2025) J Med Chem 68: 19893-19907
- PubMed: 40973081
- DOI: https://doi.org/10.1021/acs.jmedchem.4c03089
- Primary Citation of Related Structures:
8V04, 8V1F, 9E83 - PubMed Abstract:
Transmembrane protease serine-2 (TMPRSS2) is an essential host entry factor in human airways for SARS-CoV-2 and influenza A/B and has presented as a target for antiviral drug development; however, no clinically viable oral small-molecule TMPRSS2 inhibitors have been developed to date. Here, we perform two large-scale docking campaigns to identify covalent and noncovalent TMPRSS2 small-molecule inhibitors using a homology model and crystal structure. We establish a pipeline to rapidly screen TMPRSS2 inhibitors and then interrogate the potency, selectivity, and biophysical properties of covalent and noncovalent inhibition using enzyme kinetics on synthetic peptide and protein substrates and differential scanning fluorimetry. Furthermore, we established a readily crystallizable form of TMPRSS2 protein that produced high-resolution crystal structures with nafamostat , '157 , and 6-amidino-2-naphthol . A novel noncovalent inhibitor scaffold is biochemically validated as a potential avenue for developing TMPRSS2-selective inhibitors.
- Structural Genomics Consortium Toronto, Toronto, Ontario M5G 1L7, Canada.
Organizational Affiliation: