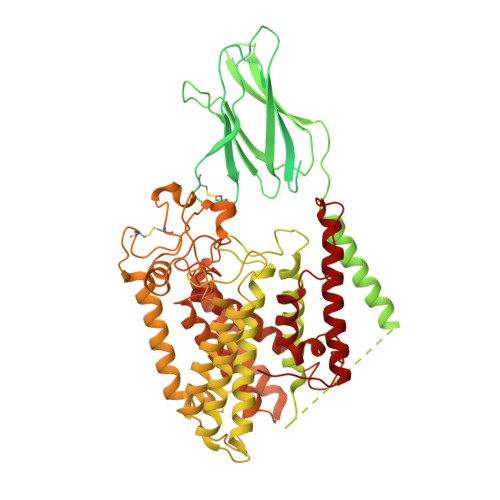
Structure of the human heparan-alpha-glucosaminide N -acetyltransferase (HGSNAT).
Navratna, V., Kumar, A., Rana, J.K., Mosalaganti, S.(2024) Elife 13
- PubMed: 39196614
- DOI: https://doi.org/10.7554/eLife.93510
- Primary Citation of Related Structures:
8TU9 - PubMed Abstract:
Degradation of heparan sulfate (HS), a glycosaminoglycan (GAG) comprised of repeating units of N -acetylglucosamine and glucuronic acid, begins in the cytosol and is completed in the lysosomes. Acetylation of the terminal non-reducing amino group of α-D-glucosamine of HS is essential for its complete breakdown into monosaccharides and free sulfate. Heparan-α-glucosaminide N -acetyltransferase (HGSNAT), a resident of the lysosomal membrane, catalyzes this essential acetylation reaction by accepting and transferring the acetyl group from cytosolic acetyl-CoA to terminal α-D-glucosamine of HS in the lysosomal lumen. Mutation-induced dysfunction in HGSNAT causes abnormal accumulation of HS within the lysosomes and leads to an autosomal recessive neurodegenerative lysosomal storage disorder called mucopolysaccharidosis IIIC (MPS IIIC). There are no approved drugs or treatment strategies to cure or manage the symptoms of, MPS IIIC. Here, we use cryo-electron microscopy (cryo-EM) to determine a high-resolution structure of the HGSNAT-acetyl-CoA complex, the first step in the HGSNAT-catalyzed acetyltransferase reaction. In addition, we map the known MPS IIIC mutations onto the structure and elucidate the molecular basis for mutation-induced HGSNAT dysfunction.
- Life Sciences Institute, University of Michigan, Ann Arbor, United States.
Organizational Affiliation: