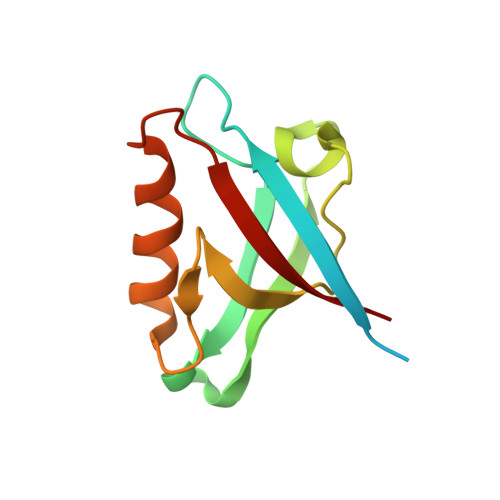
Interactions of the protein tyrosine phosphatase PTPN3 with viral and cellular partners through its PDZ domain: insights into structural determinants and phosphatase activity.
Genera, M., Colcombet-Cazenave, B., Croitoru, A., Raynal, B., Mechaly, A., Caillet, J., Haouz, A., Wolff, N., Caillet-Saguy, C.(2023) Front Mol Biosci 10: 1192621-1192621
- PubMed: 37200868
- DOI: https://doi.org/10.3389/fmolb.2023.1192621
- Primary Citation of Related Structures:
8CQY, 8OEP - PubMed Abstract:
The human protein tyrosine phosphatase non-receptor type 3 (PTPN3) is a phosphatase containing a PDZ (PSD-95/Dlg/ZO-1) domain that has been found to play both tumor-suppressive and tumor-promoting roles in various cancers, despite limited knowledge of its cellular partners and signaling functions. Notably, the high-risk genital human papillomavirus (HPV) types 16 and 18 and the hepatitis B virus (HBV) target the PDZ domain of PTPN3 through PDZ-binding motifs (PBMs) in their E6 and HBc proteins respectively. This study focuses on the interactions between the PTPN3 PDZ domain (PTPN3-PDZ) and PBMs of viral and cellular protein partners. We solved the X-ray structures of complexes between PTPN3-PDZ and PBMs of E6 of HPV18 and the tumor necrosis factor-alpha converting enzyme (TACE). We provide new insights into key structural determinants of PBM recognition by PTPN3 by screening the selectivity of PTPN3-PDZ recognition of PBMs, and by comparing the PDZome binding profiles of PTPN3-recognized PBMs and the interactome of PTPN3-PDZ. The PDZ domain of PTPN3 was known to auto-inhibit the protein's phosphatase activity. We discovered that the linker connecting the PDZ and phosphatase domains is involved in this inhibition, and that the binding of PBMs does not impact this catalytic regulation. Overall, the study sheds light on the interactions and structural determinants of PTPN3 with its cellular and viral partners, as well as on the inhibitory role of its PDZ domain on its phosphatase activity.
- Institut Pasteur, Université Paris Cité, Channel Receptors Unit, Paris, France.
Organizational Affiliation: