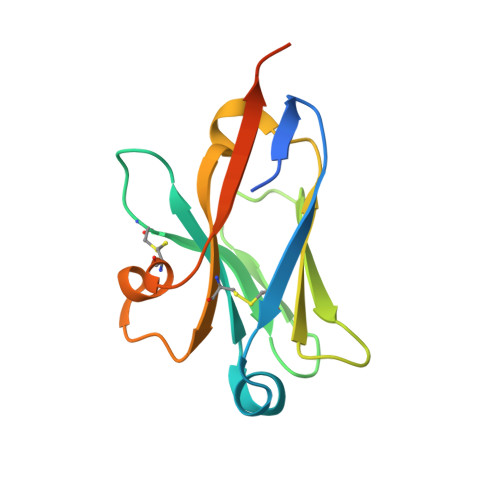
Structural insights into the unique recognition module between alpha-synuclein peptide and nanobody.
Islam, Z., Vaikath, N.N., Hmila, I., El-Agnaf, O.M.A., Kolatkar, P.R.(2024) Protein Sci 33: e4875-e4875
- PubMed: 38105512
- DOI: https://doi.org/10.1002/pro.4875
- Primary Citation of Related Structures:
8JJV, 8JLY - PubMed Abstract:
Nanobodies are single-domain fragments of antibodies with comparable specificity and affinity to antibodies. They are emerging as versatile tools in biology due to their relatively small size. Here, we report the crystal structure of a specific nanobody Nbα-syn01, bound to a 14 amino acid long peptide of α-synuclein (αSyn), a 140-residue protein whose aggregation is associated with Parkinson's disease. The complex structure exhibits a unique binding pattern where the αSyn peptide replaces the N-terminal region of nanobody. Recognition is mediated principally by extended main chain interaction of the αSyn peptide and specificity of the interaction lies in the central 48-52 region of αSyn peptide. Structure-guided truncation of Nbα-syn01 shows tighter binding to αSyn peptide and improved inhibition of α-synuclein aggregation. The structure of the truncated complex was subsequently determined and was indistinguishable to full length complex as the full-length form had no visible electron density for the N-terminal end. These findings reveal the molecular basis for a previously unobserved binding mode for nanobody recognition of α-synuclein, providing an explanation for the enhanced binding, and potential for an alternate framework for structure-based protein engineering of nanobodies to develop better diagnostic and therapeutic tools.
- Diabetes Center, Qatar Biomedical Research Institute, Hamad Bin Khalifa University, Qatar Foundation, Doha, Qatar.
Organizational Affiliation: