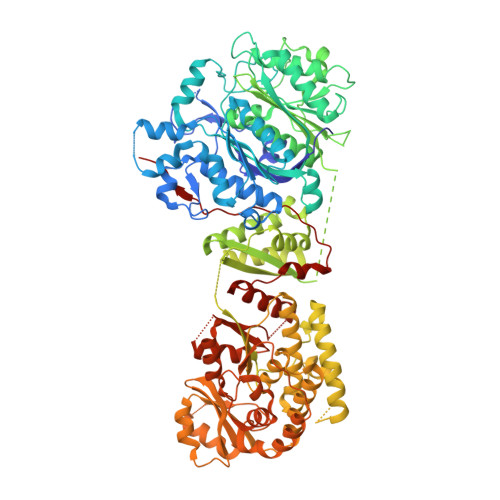
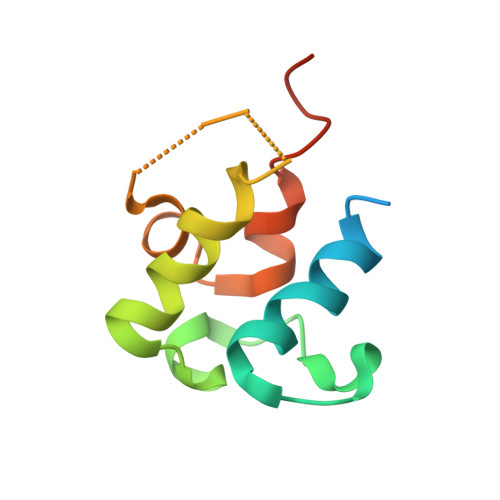
Structure-Based Analysis of Transient Interactions between Ketosynthase-like Decarboxylase and Acyl Carrier Protein in a Loading Module of Modular Polyketide Synthase.
Chisuga, T., Murakami, S., Miyanaga, A., Kudo, F., Eguchi, T.(2023) ACS Chem Biol 18: 1398-1404
- PubMed: 37216195
- DOI: https://doi.org/10.1021/acschembio.3c00151
- Primary Citation of Related Structures:
8IN9 - PubMed Abstract:
Ketosynthase-like decarboxylase (KS Q ) domains are widely distributed in the loading modules of modular type I polyketide synthases (PKSs) and catalyze the decarboxylation of the (alkyl-)malonyl unit bound to the acyl carrier protein (ACP) in the loading module for the construction of the PKS starter unit. Previously, we performed a structural and functional analysis of the GfsA KS Q domain involved in the biosynthesis of macrolide antibiotic FD-891. We furthermore revealed the recognition mechanism for the malonic acid thioester moiety of the malonyl-GfsA loading module ACP (ACP L ) as a substrate. However, the exact recognition mechanism for the GfsA ACP L moiety remains unclear. Here, we present a structural basis for the interactions between the GfsA KS Q domain and GfsA ACP L . We determined the crystal structure of the GfsA KS Q -acyltransferase (AT) didomain in complex with ACP L (ACP L =KS Q AT complex) by using a pantetheine crosslinking probe. We identified the key amino acid residues involved in the KS Q domain-ACP L interactions and confirmed the importance of these residues by mutational analysis. The binding mode of ACP L to the GfsA KS Q domain is similar to that of ACP to the ketosynthase domain in modular type I PKSs. Furthermore, comparing the ACP L =KS Q AT complex structure with other full-length PKS module structures provides important insights into the overall architectures and conformational dynamics of the type I PKS modules.
- Department of Chemistry, Tokyo Institute of Technology, 2-12-1 O-Okayama, Tokyo 152-8551, Japan.
Organizational Affiliation: