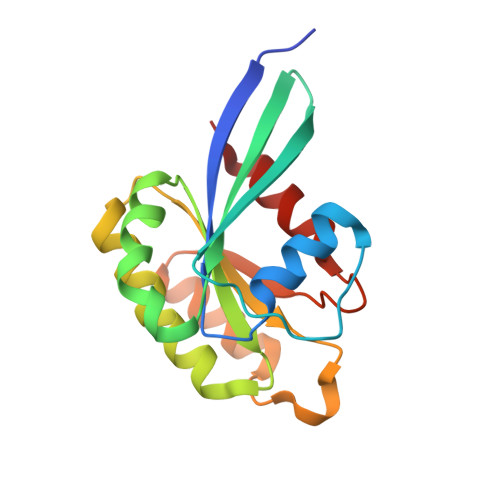
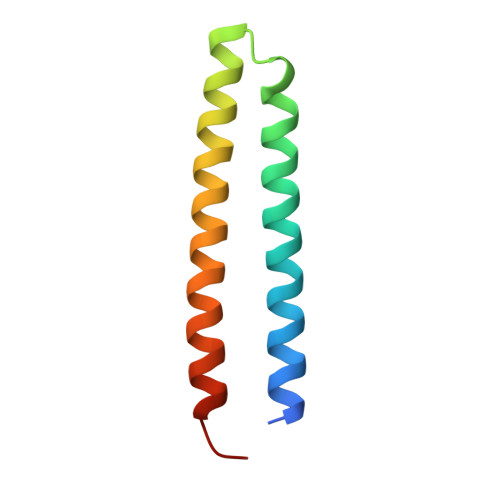
Structural basis of ELKS/Rab6B interaction and its role in vesicle capturing enhanced by liquid-liquid phase separation.
Jin, G., Lin, L., Li, K., Li, J., Yu, C., Wei, Z.(2023) J Biological Chem 299: 104808-104808
- PubMed: 37172719
- DOI: https://doi.org/10.1016/j.jbc.2023.104808
- Primary Citation of Related Structures:
8IJ9 - PubMed Abstract:
ELKS proteins play a key role in organizing intracellular vesicle trafficking and targeting in both neurons and non-neuronal cells. While it is known that ELKS interacts with the vesicular traffic regulator, the Rab6 GTPase, the molecular basis governing ELKS-mediated trafficking of Rab6-coated vesicles, has remained unclear. In this study, we solved the Rab6B structure in complex with the Rab6-binding domain of ELKS1, revealing that a C-terminal segment of ELKS1 forms a helical hairpin to recognize Rab6B through a unique binding mode. We further showed that liquid-liquid phase separation (LLPS) of ELKS1 allows it to compete with other Rab6 effectors for binding to Rab6B and accumulate Rab6B-coated liposomes to the protein condensate formed by ELKS1. We also found that the ELKS1 condensate recruits Rab6B-coated vesicles to vesicle-releasing sites and promotes vesicle exocytosis. Together, our structural, biochemical, and cellular analyses suggest that ELKS1, via the LLPS-enhanced interaction with Rab6, captures Rab6-coated vesicles from the cargo transport machine for efficient vesicle release at exocytotic sites. These findings shed new light on the understanding of spatiotemporal regulation of vesicle trafficking through the interplay between membranous structures and membraneless condensates.
- Brain Research Center, Southern University of Science and Technology, Shenzhen, Guangdong, China; School of Life Sciences, Southern University of Science and Technology, Shenzhen, Guangdong, China.
Organizational Affiliation: