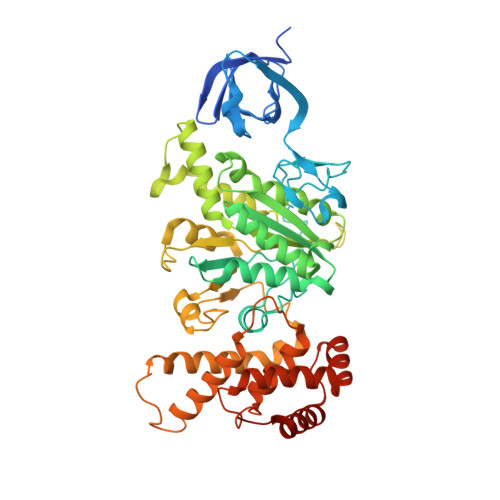
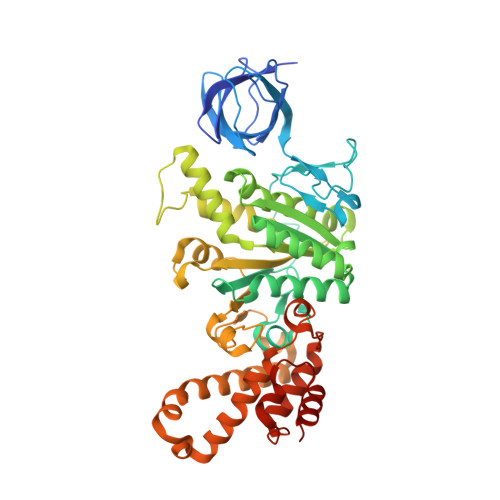
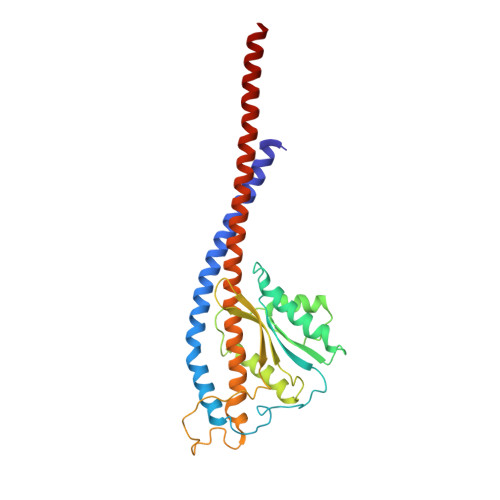
Mechanism of ATP hydrolysis dependent rotation of bacterial ATP synthase.
Nakano, A., Kishikawa, J.I., Mitsuoka, K., Yokoyama, K.(2023) Nat Commun 14: 4090-4090
- PubMed: 37429854
- DOI: https://doi.org/10.1038/s41467-023-39742-5
- Primary Citation of Related Structures:
8HH1, 8HH2, 8HH3, 8HH4, 8HH5, 8HH6, 8HH7, 8HH8, 8HH9, 8HHA, 8HHB, 8HHC - PubMed Abstract:
F 1 domain of ATP synthase is a rotary ATPase complex in which rotation of central γ-subunit proceeds in 120° steps against a surrounding α 3 β 3 fueled by ATP hydrolysis. How the ATP hydrolysis reactions occurring in three catalytic αβ dimers are coupled to mechanical rotation is a key outstanding question. Here we describe catalytic intermediates of the F 1 domain in F o F 1 synthase from Bacillus PS3 sp. during ATP mediated rotation captured using cryo-EM. The structures reveal that three catalytic events and the first 80° rotation occur simultaneously in F 1 domain when nucleotides are bound at all the three catalytic αβ dimers. The remaining 40° rotation of the complete 120° step is driven by completion of ATP hydrolysis at α D β D , and proceeds through three sub-steps (83°, 91°, 101°, and 120°) with three associated conformational intermediates. All sub-steps except for one between 91° and 101° associated with phosphate release, occur independently of the chemical cycle, suggesting that the 40° rotation is largely driven by release of intramolecular strain accumulated by the 80° rotation. Together with our previous results, these findings provide the molecular basis of ATP driven rotation of ATP synthases.
- Department of Molecular Biosciences, Kyoto Sangyo University, Kamigamo-Motoyama, Kita-ku, Kyoto, 603-8555, Japan.
Organizational Affiliation: