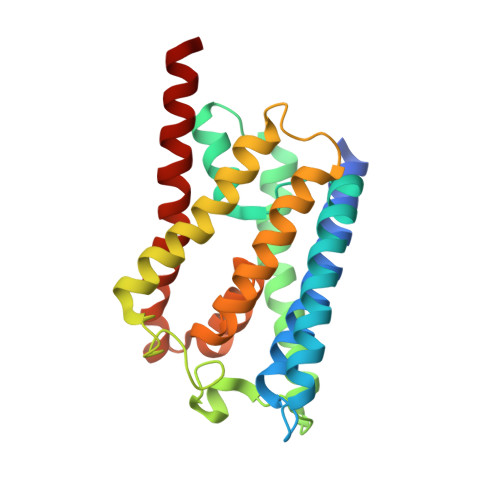
Solid-state NMR structure determination of a membrane protein in E. coli cellular inner membrane.
Xie, H., Zhao, Y., Zhao, W., Chen, Y., Liu, M., Yang, J.(2023) Sci Adv 9: eadh4168-eadh4168
- PubMed: 37910616
- DOI: https://doi.org/10.1126/sciadv.adh4168
- Primary Citation of Related Structures:
8H1D - PubMed Abstract:
Structure determination of membrane proteins in native cellular membranes is critical to precisely reveal their structures in physiological conditions. However, it remains challenging for solid-state nuclear magnetic resonance (ssNMR) due to the low sensitivity and high complexity of ssNMR spectra of cellular membranes. Here, we present the structure determination of aquaporin Z (AqpZ) by ssNMR in Escherichia coli inner membranes. To enhance the signal sensitivity of AqpZ, we optimized protein overexpression and removed outer membrane components. To suppress the interference of background proteins, we used a "dual-media" expression approach and antibiotic treatment. Using 1017 distance restraints obtained from two-dimensional 13 C- 13 C spectra based on the complete chemical shift assignments, the 1.7-Å ssNMR structure of AqpZ is determined in E. coli inner membranes. This cellular ssNMR structure determination paves the way for analyzing the atomic structural details for membrane proteins in native cellular membranes.
- National Center for Magnetic Resonance in Wuhan, Key Laboratory of Magnetic Resonance in Biological Systems, State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics, Wuhan Institute of Physics and Mathematics, Innovation Academy for Precision Measurement Science and Technology, Chinese Academy of Sciences, Wuhan 430071, P. R. China.
Organizational Affiliation: