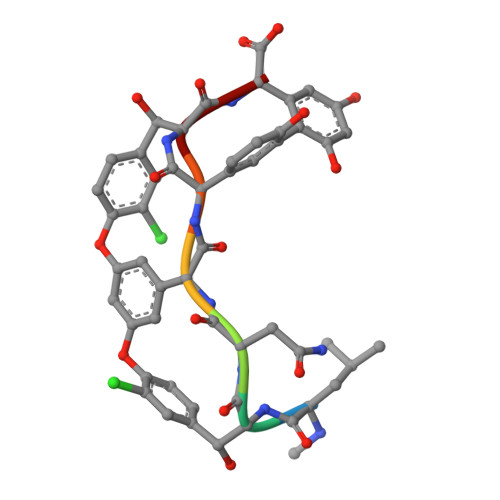
Crystal structure of vancomycin bound to the resistance determinant D-alanine-D-serine.
Park, J.H., Reviello, R.E., Loll, P.J.(2024) IUCrJ 11: 133-139
- PubMed: 38277167
- DOI: https://doi.org/10.1107/S2052252524000289
- Primary Citation of Related Structures:
8G82 - PubMed Abstract:
Vancomycin is a glycopeptide antibiotic that for decades has been a mainstay of treatment for persistent bacterial infections. However, the spread of antibiotic resistance threatens its continued utility. In particular, vancomycin-resistant enterococci (VRE) have become a pressing clinical challenge. Vancomycin acts by binding and sequestering the intermediate Lipid II in cell-wall biosynthesis, specifically recognizing a D-alanine-D-alanine dipeptide motif within the Lipid II molecule. VRE achieve resistance by remodeling this motif to either D-alanine-D-lactate or D-alanine-D-serine; the former substitution essentially abolishes recognition by vancomycin of Lipid II, whereas the latter reduces the affinity of the antibiotic by roughly one order of magnitude. The complex of vancomycin bound to D-alanine-D-serine has been crystallized, and its 1.20 Å X-ray crystal structure is presented here. This structure reveals that the D-alanine-D-serine ligand is bound in essentially the same position and same pose as the native D-alanine-D-alanine ligand. The serine-containing ligand appears to be slightly too large to be comfortably accommodated in this way, suggesting one possible contribution to the reduced binding affinity. In addition, two flexible hydroxyl groups - one from the serine side chain of the ligand, and the other from a glucose sugar on the antibiotic - are locked into single conformations in the complex, which is likely to contribute an unfavorable entropic component to the recognition of the serine-containing ligand.
- Department of Biochemistry and Molecular Biology, Drexel University College of Medicine, PA 19102, USA.
Organizational Affiliation: