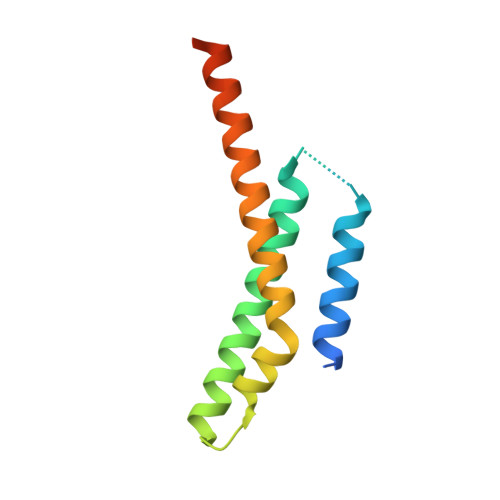
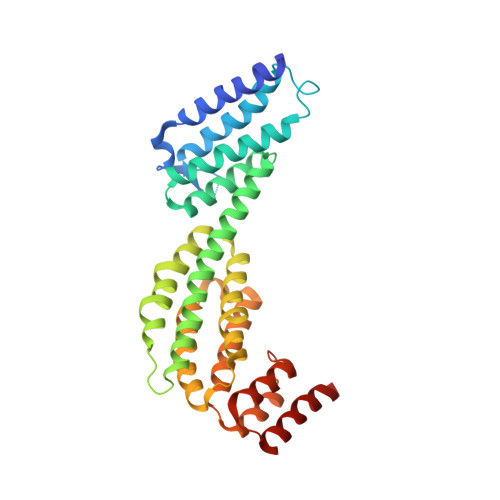
Structure of a membrane tethering complex incorporating multiple SNAREs.
DAmico, K.A., Stanton, A.E., Shirkey, J.D., Travis, S.M., Jeffrey, P.D., Hughson, F.M.(2024) Nat Struct Mol Biol 31: 246-254
- PubMed: 38196032
- DOI: https://doi.org/10.1038/s41594-023-01164-8
- Primary Citation of Related Structures:
8EKI, 8FTU - PubMed Abstract:
Most membrane fusion reactions in eukaryotic cells are mediated by multisubunit tethering complexes (MTCs) and SNARE proteins. MTCs are much larger than SNAREs and are thought to mediate the initial attachment of two membranes. Complementary SNAREs then form membrane-bridging complexes whose assembly draws the membranes together for fusion. Here we present a cryo-electron microscopy structure of the simplest known MTC, the 255-kDa Dsl1 complex of Saccharomyces cerevisiae, bound to the two SNAREs that anchor it to the endoplasmic reticulum. N-terminal domains of the SNAREs form an integral part of the structure, stabilizing a Dsl1 complex configuration with unexpected similarities to the 850-kDa exocyst MTC. The structure of the SNARE-anchored Dsl1 complex and its comparison with exocyst reveal what are likely to be common principles underlying MTC function. Our structure also implies that tethers and SNAREs can work together as a single integrated machine.
- Department of Molecular Biology, Princeton University, Princeton, NJ, USA.
Organizational Affiliation: