Antiviral Activity and Crystal Structures of HIV-1 gp120 Antagonists.
Curreli, F., Kwon, Y.D., Nicolau, I., Burgos, G., Altieri, A., Kurkin, A.V., Verardi, R., Kwong, P.D., Debnath, A.K.(2022) Int J Mol Sci 23
- PubMed: 36555641
- DOI: https://doi.org/10.3390/ijms232415999
- Primary Citation of Related Structures:
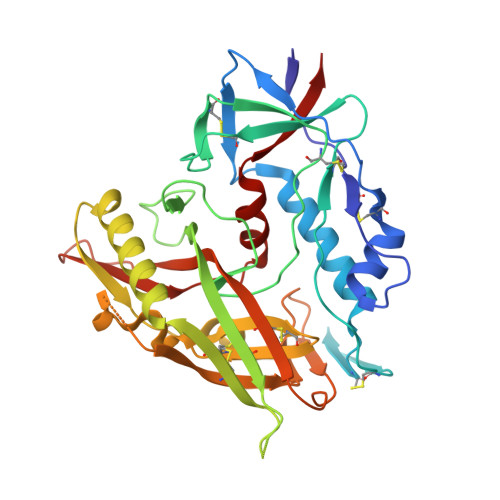
8F9Z, 8FA0 - PubMed Abstract:
As part of our effort to discover drugs that target HIV-1 entry, we report the antiviral activity and crystal structures of two novel inhibitors in a complex with a gp120 core. NBD-14204 showed similar antiviral activity against all the clinical isolates tested. The IC 50 values were in the range of 0.24-0.9 µM with an overall mean of 0.47 ± 0.03 µM, showing slightly better activity against the clinical isolates than against the lab-adapted HIV-1 HXB2 (IC 50 = 0.96 ± 0.1 µM) . Moreover, the antiviral activity of NBD-14208 was less consistent, showing a wider range of IC 50 values (0.66-5.7 µM) with an overall mean of 3 ± 0.25 µM and better activity against subtypes B and D (Mean IC 50 2.2-2.5 µM) than the A, C and Rec viruses (Mean IC 50 2.9-3.9 µM). SI of NBD-14204 was about 10-fold higher than NBD-14208, making it a better lead compound for further optimization. In addition, we tested these compounds against S375Y and S375H mutants of gp120, which occurred in some clades and observed these to be sensitive to NBD-14204 and NBD-14208. These inhibitors also showed modest activity against HIV-1 reverse transcriptase. Furthermore, we determined the crystal structures of both inhibitors in complexes with gp120 cores. As expected, both NBD-14204 and NBD-14208 bind primarily within the Phe43 cavity. It is noteworthy that the electron density of the thiazole ring in both structures was poorly defined due to the flexibility of this scaffold, suggesting that these compounds maintain substantial entropy, even when bound to the Phe43 cavity.
- Laboratory of Molecular Modeling and Drug Design, Lindsey F. Kimball Research Institute, New York Blood Center, New York, NY 10065, USA.
Organizational Affiliation: