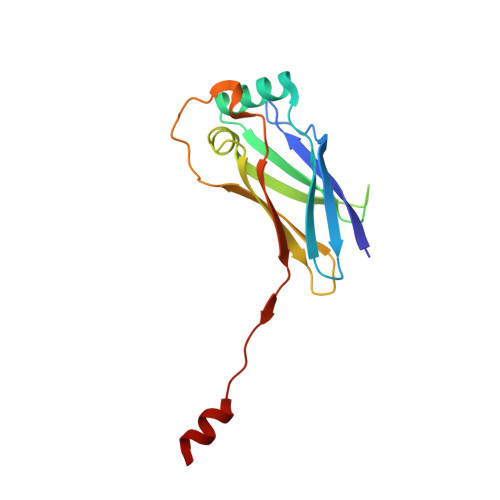
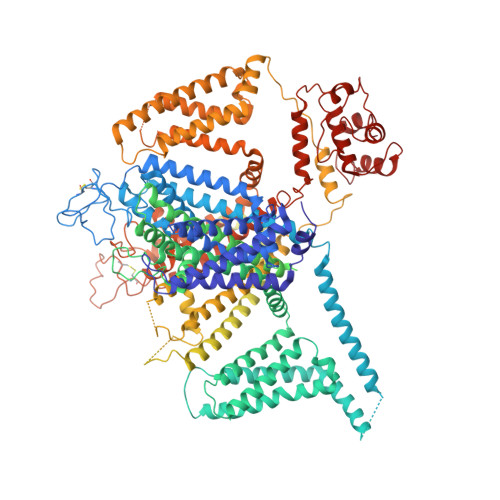
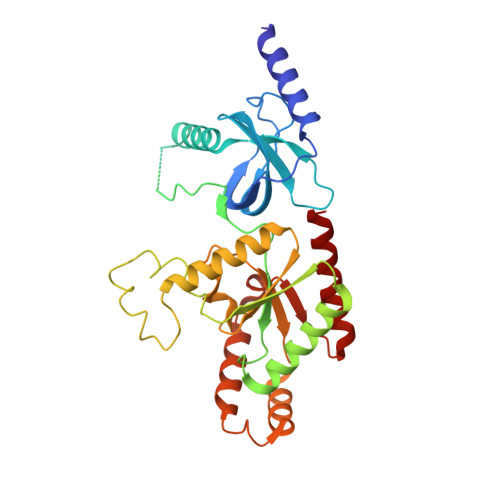
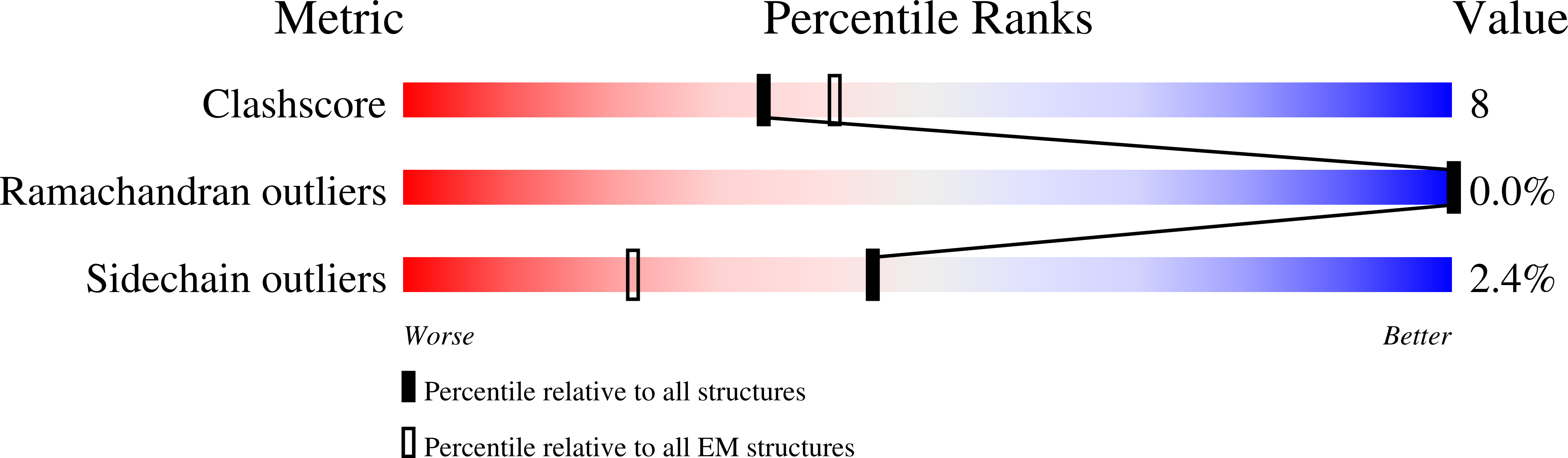EMC chaperone-Ca V structure reveals an ion channel assembly intermediate.
Chen, Z., Mondal, A., Abderemane-Ali, F., Jang, S., Niranjan, S., Montano, J.L., Zaro, B.W., Minor Jr., D.L.(2023) Nature 619: 410-419
- PubMed: 37196677
- DOI: https://doi.org/10.1038/s41586-023-06175-5
- Primary Citation of Related Structures:
8EOG, 8EOI - PubMed Abstract:
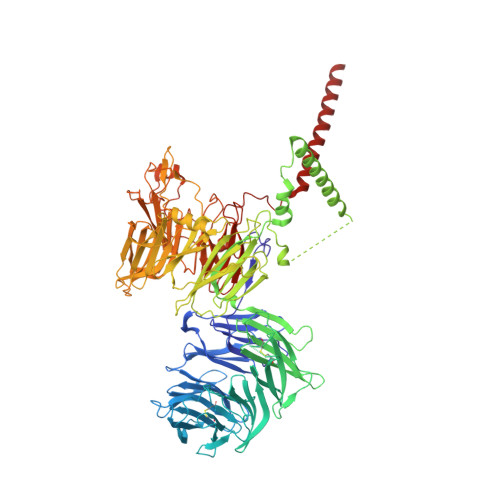
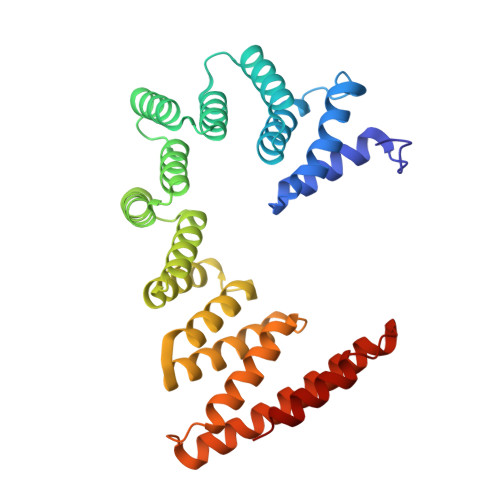
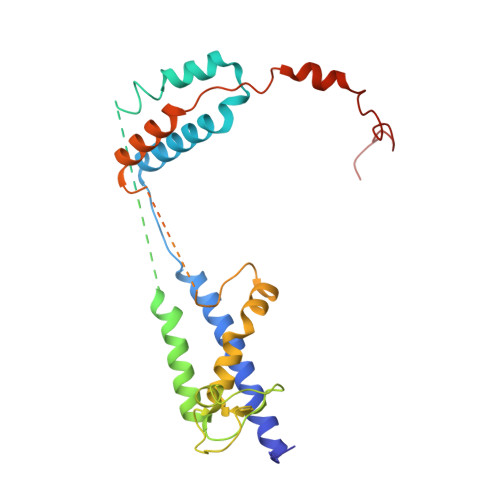
Voltage-gated ion channels (VGICs) comprise multiple structural units, the assembly of which is required for function 1,2 . Structural understanding of how VGIC subunits assemble and whether chaperone proteins are required is lacking. High-voltage-activated calcium channels (Ca V s) 3,4 are paradigmatic multisubunit VGICs whose function and trafficking are powerfully shaped by interactions between pore-forming Ca V 1 or Ca V 2 Ca V α 1 (ref. 3 ), and the auxiliary Ca V β 5 and Ca V α 2 δ subunits 6,7 . Here we present cryo-electron microscopy structures of human brain and cardiac Ca V 1.2 bound with Ca V β 3 to a chaperone-the endoplasmic reticulum membrane protein complex (EMC) 8,9 -and of the assembled Ca V 1.2-Ca V β 3 -Ca V α 2 δ-1 channel. These structures provide a view of an EMC-client complex and define EMC sites-the transmembrane (TM) and cytoplasmic (Cyto) docks; interaction between these sites and the client channel causes partial extraction of a pore subunit and splays open the Ca V α 2 δ-interaction site. The structures identify the Ca V α 2 δ-binding site for gabapentinoid anti-pain and anti-anxiety drugs 6 , show that EMC and Ca V α 2 δ interactions with the channel are mutually exclusive, and indicate that EMC-to-Ca V α 2 δ hand-off involves a divalent ion-dependent step and Ca V 1.2 element ordering. Disruption of the EMC-Ca V complex compromises Ca V function, suggesting that the EMC functions as a channel holdase that facilitates channel assembly. Together, the structures reveal a Ca V assembly intermediate and EMC client-binding sites that could have wide-ranging implications for the biogenesis of VGICs and other membrane proteins.
- Cardiovascular Research Institute, University of California, San Francisco, CA, USA.
Organizational Affiliation: