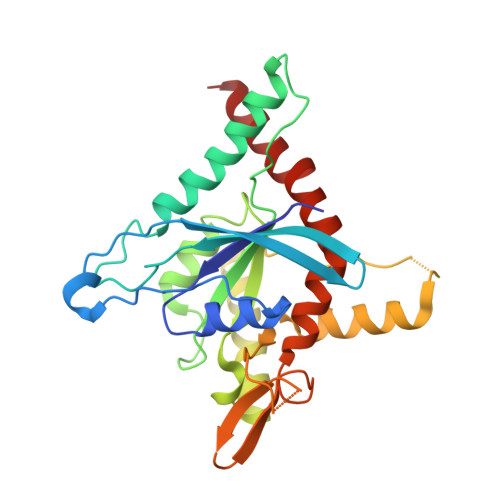
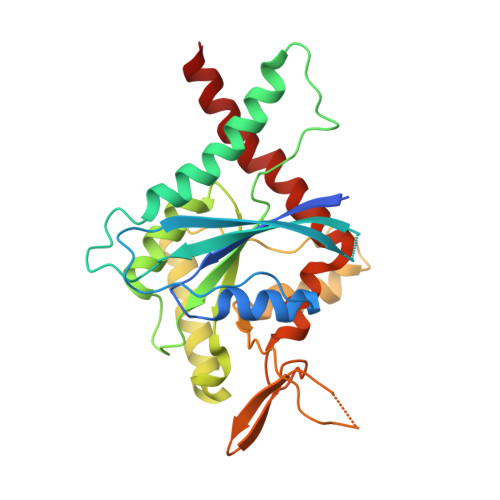
Conservation and divergence of the G-interfaces of Drosophila melanogaster septins.
de Freitas Fernandes, A., Leonardo, D.A., Cavini, I.A., Rosa, H.V.D., Vargas, J.A., D'Muniz Pereira, H., Nascimento, A.S., Garratt, R.C.(2023) Cytoskeleton (Hoboken) 80: 153-168
- PubMed: 36576069
- DOI: https://doi.org/10.1002/cm.21740
- Primary Citation of Related Structures:
8DKT - PubMed Abstract:
Septins possess a conserved guanine nucleotide-binding (G) domain that participates in the stabilization of organized hetero-oligomeric complexes which assemble into filaments, rings and network-like structures. The fruit fly, Drosophila melanogaster, has five such septin genes encoding Sep1, Sep2, Sep4, Sep5 and Pnut. Here, we report the crystal structure of the heterodimer formed between the G-domains of Sep1 and Sep2, the first from an insect to be described to date. A G-interface stabilizes the dimer (in agreement with the expected arrangement for the Drosophila hexameric particle) and this bears significant resemblance to its human counterparts, even down to the level of individual amino acid interactions. On the other hand, a model for the G-interface formed between the two copies of Pnut which occupy the centre of the hexamer, shows important structural differences, including the loss of a highly favourable bifurcated salt-bridge network. Whereas wild-type Pnut purifies as a monomer, the reintroduction of the salt-bridge network results in stabilizing the dimeric interface in solution as shown by size exclusion chromatography and thermal stability measurements. Adaptive steered molecular dynamics reveals an unzipping mechanism for dimer dissociation which initiates at a point of electrostatic repulsion within the switch II region. Overall, the data contribute to a better understanding of the molecular interactions involved in septin assembly/disassembly.
- São Carlos Institute of Physics, University of São Paulo, São Carlos, Brazil.
Organizational Affiliation: