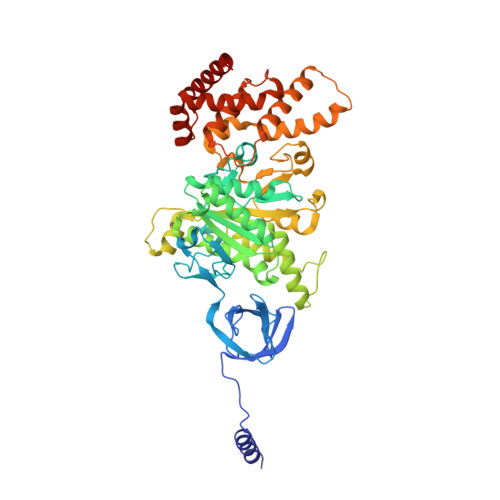
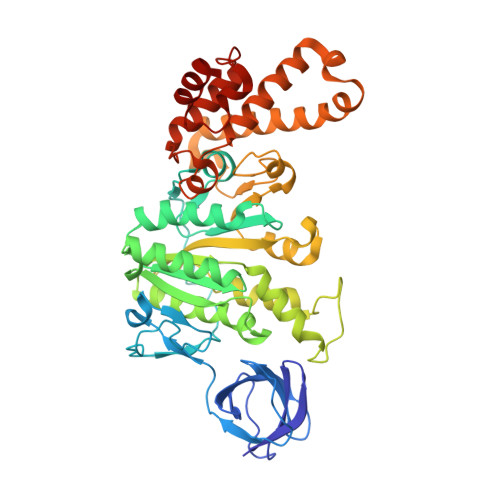
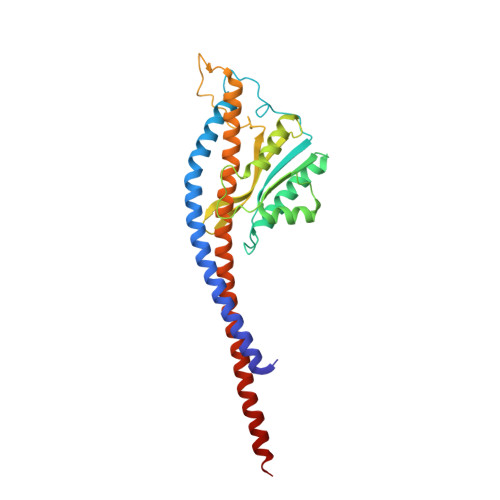
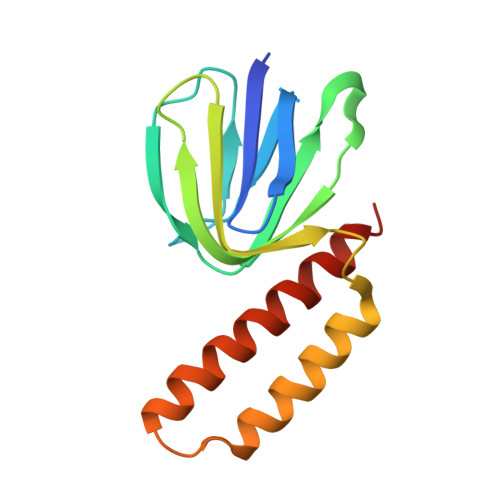
Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Sobti, M., Zeng, Y.C., Walshe, J.L., Brown, S.H.J., Ishmukhametov, R., Stewart, A.G.(2023) Commun Biol 6: 26-26
- PubMed: 36631659
- DOI: https://doi.org/10.1038/s42003-023-04414-z
- Primary Citation of Related Structures:
8DBP, 8DBQ, 8DBR, 8DBS, 8DBT, 8DBU, 8DBV, 8DBW - PubMed Abstract:
F 1 F o ATP synthase functions as a biological generator and makes a major contribution to cellular energy production. Proton flow generates rotation in the F o motor that is transferred to the F 1 motor to catalyze ATP production, with flexible F 1 /F o coupling required for efficient catalysis. F 1 F o ATP synthase can also operate in reverse, hydrolyzing ATP and pumping protons, and in bacteria this function can be regulated by an inhibitory ε subunit. Here we present cryo-EM data showing E. coli F 1 F o ATP synthase in different rotational and inhibited sub-states, observed following incubation with 10 mM MgATP. Our structures demonstrate how structural transitions within the inhibitory ε subunit induce torsional movement in the central stalk, thereby enabling its rotation within the F ο motor. This highlights the importance of the central rotor for flexible coupling of the F 1 and F o motors and provides further insight into the regulatory mechanism mediated by subunit ε.
- Molecular, Structural and Computational Biology Division, The Victor Chang Cardiac Research Institute, Darlinghurst, NSW, Australia.
Organizational Affiliation: