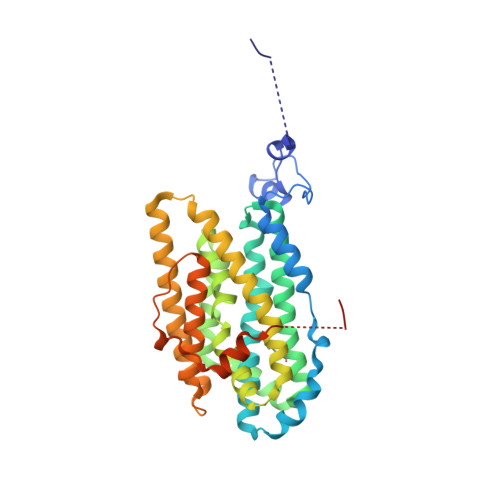
Structure of a ribonucleotide reductase R2 protein radical.
Lebrette, H., Srinivas, V., John, J., Aurelius, O., Kumar, R., Lundin, D., Brewster, A.S., Bhowmick, A., Sirohiwal, A., Kim, I.S., Gul, S., Pham, C., Sutherlin, K.D., Simon, P., Butryn, A., Aller, P., Orville, A.M., Fuller, F.D., Alonso-Mori, R., Batyuk, A., Sauter, N.K., Yachandra, V.K., Yano, J., Kaila, V.R.I., Sjoberg, B.M., Kern, J., Roos, K., Hogbom, M.(2023) Science 382: 109-113
- PubMed: 37797025
- DOI: https://doi.org/10.1126/science.adh8160
- Primary Citation of Related Structures:
8BT3, 8BT4 - PubMed Abstract:
Aerobic ribonucleotide reductases (RNRs) initiate synthesis of DNA building blocks by generating a free radical within the R2 subunit; the radical is subsequently shuttled to the catalytic R1 subunit through proton-coupled electron transfer (PCET). We present a high-resolution room temperature structure of the class Ie R2 protein radical captured by x-ray free electron laser serial femtosecond crystallography. The structure reveals conformational reorganization to shield the radical and connect it to the translocation path, with structural changes propagating to the surface where the protein interacts with the catalytic R1 subunit. Restructuring of the hydrogen bond network, including a notably short O-O interaction of 2.41 angstroms, likely tunes and gates the radical during PCET. These structural results help explain radical handling and mobilization in RNR and have general implications for radical transfer in proteins.
- Department of Biochemistry and Biophysics, Stockholm University, Arrhenius Laboratories for Natural Sciences, Stockholm, Sweden.
Organizational Affiliation: